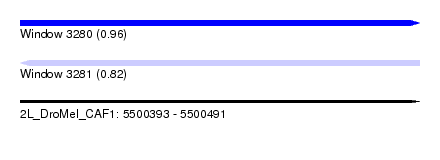
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,500,393 – 5,500,491 |
| Length | 98 |
| Max. P | 0.963657 |

| Location | 5,500,393 – 5,500,491 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
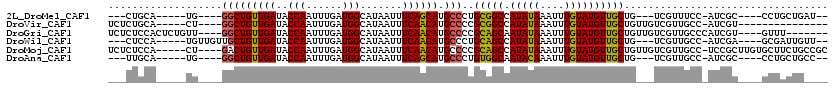
>2L_DroMel_CAF1 5500393 98 + 22407834 --AUCAGCAGG----GCGAU-GGAAACGA---CAGCAACAUACAAAUUUAUAUGGCCGCAGGGCAUGCUGAAAUUAUGCCAUCAAAUUGGUAUCAACAGCC----CA-----UGCAG--- --....(((((----((..(-(....)).---(((((.((((........))))(((....))).))))).....((((((......)))))).....)))----).-----)))..--- ( -32.40) >DroVir_CAF1 174621 95 + 1 ---------------ACGAU-GGCAACGACAACAGCAACAUACAAAUUUAUAUGGCCGCGGGGCAUGUUGAAAUUAUGCCAUCAAAUUGGUAUCAACAGCC----AG-----UGCAGAGA ---------------..(((-((((.......(((((.((((........))))(((....))).)))))......)))))))..((((((.......)))----))-----)....... ( -25.92) >DroGri_CAF1 185107 105 + 1 -------AAAC----ACGAUGGGCAACGACAACAGCAACAUACAAAUUUAUAUUGCUGCGGGGCAUGUUGAAAUUAUGCCAUCAAAUUGGUAUCAACAGCC----AACAGAGUGGAGAGA -------...(----((..(((....)..(..((((((.(((......))).))))))..)(((.((((((.....(((((......))))))))))))))----..))..)))...... ( -30.80) >DroWil_CAF1 176710 102 + 1 --AACAAUCGC----UCGAU-GGCAACGA---CAGCAACAUACAAAUUUAUAUGGCUGCAGGGCAUGUUGAAAUUAUGCCAUCAAAUUGGUAUCAACAGCAACAACA-----UGGAG--- --........(----((.((-(....(..---((((..((((........))))))))..).((.((((((.....(((((......))))))))))))).....))-----).)))--- ( -24.80) >DroMoj_CAF1 192205 110 + 1 GCGGCAGAAGCACAAGCGGA-GGCAACGACAACAGCAACAUACAAAUUUAUAUGGCUGCGGGGCAUGUUGAAAUUAUGCCAUCAAAUUGGUAUCAACAGUC----AG-----UGGAGAGA (((((....((....))(..-(....)..)........((((........)))))))))..(((.((((((.....(((((......))))))))))))))----..-----........ ( -27.00) >DroAna_CAF1 129515 98 + 1 --GGCAGCAGG----GCGAU-GGCAACGA---CAGCAACAUACAAAUUUGUAUUGCCACAGGGCAUGCUGAAAUUAUGCCAUCAAAUUGGUAUCAACAGCC----CA-----UGCAA--- --....(((((----(((((-((((....---(((((..(((((....)))))((((....)))))))))......)))))))....((.......)))))----).-----)))..--- ( -36.20) >consensus ____CAGAAGC____GCGAU_GGCAACGA___CAGCAACAUACAAAUUUAUAUGGCCGCAGGGCAUGUUGAAAUUAUGCCAUCAAAUUGGUAUCAACAGCC____AA_____UGCAG___ .....................(((........(((((.((((........))))(((....))).))))).....((((((......)))))).....)))................... (-15.53 = -15.58 + 0.06)

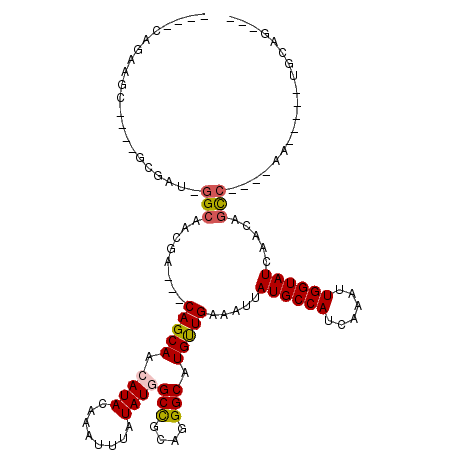
| Location | 5,500,393 – 5,500,491 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.25 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5500393 98 - 22407834 ---CUGCA-----UG----GGCUGUUGAUACCAAUUUGAUGGCAUAAUUUCAGCAUGCCCUGCGGCCAUAUAAAUUUGUAUGUUGCUG---UCGUUUCC-AUCGC----CCUGCUGAU-- ---..(((-----.(----(((.((((....))))..(((((...(((..(((((.(((....)))((((((....)))))).)))))---..))).))-)))))----)))))....-- ( -31.80) >DroVir_CAF1 174621 95 - 1 UCUCUGCA-----CU----GGCUGUUGAUACCAAUUUGAUGGCAUAAUUUCAACAUGCCCCGCGGCCAUAUAAAUUUGUAUGUUGCUGUUGUCGUUGCC-AUCGU--------------- ..((.(((-----..----...))).)).........(((((((......(((((.(((....)))((((((....))))))....)))))....))))-)))..--------------- ( -22.20) >DroGri_CAF1 185107 105 - 1 UCUCUCCACUCUGUU----GGCUGUUGAUACCAAUUUGAUGGCAUAAUUUCAACAUGCCCCGCAGCAAUAUAAAUUUGUAUGUUGCUGUUGUCGUUGCCCAUCGU----GUUU------- ......(((..((..----(((...(((((..........(((((.........)))))..(((((((((((......))))))))))))))))..)))))..))----)...------- ( -26.90) >DroWil_CAF1 176710 102 - 1 ---CUCCA-----UGUUGUUGCUGUUGAUACCAAUUUGAUGGCAUAAUUUCAACAUGCCCUGCAGCCAUAUAAAUUUGUAUGUUGCUG---UCGUUGCC-AUCGA----GCGAUUGUU-- ---...((-----(((((.((((((..(.......)..))))))......)))))))....(((((((((((....))))))..))))---).(((((.-.....----)))))....-- ( -25.70) >DroMoj_CAF1 192205 110 - 1 UCUCUCCA-----CU----GACUGUUGAUACCAAUUUGAUGGCAUAAUUUCAACAUGCCCCGCAGCCAUAUAAAUUUGUAUGUUGCUGUUGUCGUUGCC-UCCGCUUGUGCUUCUGCCGC ........-----..----(((((((((..(((......))).......))))))......(((((((((((....))))))..))))).)))((.((.-...((....))....)).)) ( -19.60) >DroAna_CAF1 129515 98 - 1 ---UUGCA-----UG----GGCUGUUGAUACCAAUUUGAUGGCAUAAUUUCAGCAUGCCCUGUGGCAAUACAAAUUUGUAUGUUGCUG---UCGUUGCC-AUCGC----CCUGCUGCC-- ---..(((-----.(----(((.((((....))))..(((((((......(((((((((....))))(((((....)))))..)))))---....))))-)))))----)))))....-- ( -37.20) >consensus ___CUCCA_____UG____GGCUGUUGAUACCAAUUUGAUGGCAUAAUUUCAACAUGCCCCGCAGCCAUAUAAAUUUGUAUGUUGCUG___UCGUUGCC_AUCGC____GCUGCUG____ ...................(((((((((..(((......))).......)))))).)))..(((((.(((((....)))))))))).................................. (-16.67 = -16.25 + -0.42)
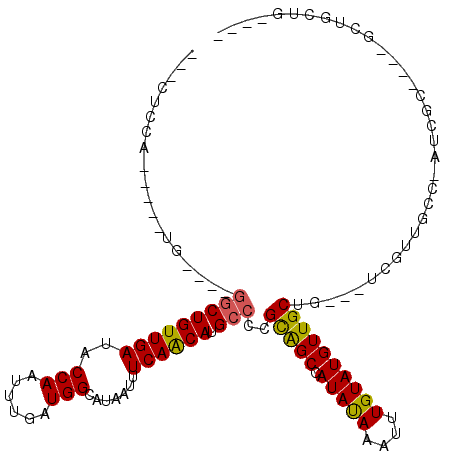
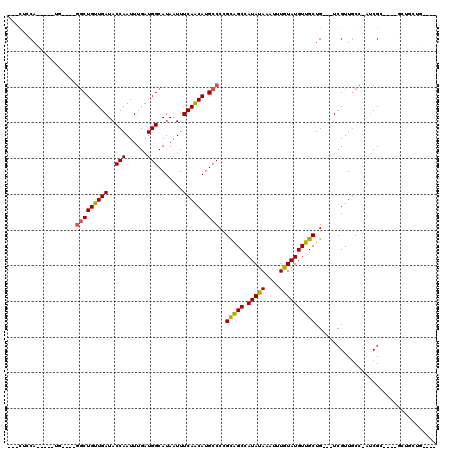
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:45 2006