| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,497,343 – 5,497,439 |
| Length | 96 |
| Max. P | 0.666779 |

| Location | 5,497,343 – 5,497,439 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.55 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5497343 96 + 22407834 AUAUGUAUAAGCUGAUUAGGAGAAGGAAAAGUGCUCGAGAG-CCAUAAACCUUUGCCAAAGACUCAACUUUUGUUAGGACUAAGCAGCUUAUAUCAG ....((((((((((.(((((..((((......(((....))-)......))))..)).....(..(((....)))..)..))).))))))))))... ( -24.40) >DroPse_CAF1 136912 81 + 1 ---UGCGUAAGCUGAUUAGG-------------CUUCAGAGACCUCAGACCUCUGCAACAGCCCCAACUUUUGUUUAGUUUAAGCUGCUUAUAGCAG ---(((((((((...(((((-------------((.(((((.........)))))....))))..((((.......)))))))...)))))).))). ( -19.30) >DroSec_CAF1 121128 96 + 1 GUAUAUAUAAGCUGAUUAGGAGAAGGAAAAGUGCUCAAAAG-CCAUAAACCUCUGCCAAAGACUCAACUUUUGUUAGGACUAAGCAGCUUAUAUCAG ....((((((((((.((((...........(.(((....))-)).....(((..((.((((......)))).)).))).)))).))))))))))... ( -22.30) >DroSim_CAF1 127926 96 + 1 GUAUAUAUAAGCUGAUUAGGAGAAGGAAAAGUGCUCGAGAG-CCAUAAACCUUUGCCAAAGACUUAACUUUUGUUAGGACUAAGCAGCUUAUAUCAG ....((((((((((.(((((..((((......(((....))-)......))))..)).....((((((....))))))..))).))))))))))... ( -27.40) >DroEre_CAF1 118838 93 + 1 GUAUAUAUAAGCUGAUUAGGAGAGGAAAAAGUGCUCGAGAG-CCAUAAACCUGUGCAGCGGACUC---UUUUGUUAGGACUAAGCAGCUUAUAUCAG ....((((((((((.((((.......(((((.(..((...(-(((......)).))..))..).)---)))).......)))).))))))))))... ( -24.74) >DroYak_CAF1 125606 96 + 1 GUAUAUAUAAGCUGAUUAGGAGAAGAAAAAGUGCUCGAGAG-CCAUAAACCUGUGCAAGAGACUCAACUUUUGUUAGGACUAAGCAGCUUAUAUCAG ....((((((((((.((((...........(.(((....))-)).....((((.(((((((......))))))))))).)))).))))))))))... ( -28.00) >consensus GUAUAUAUAAGCUGAUUAGGAGAAGGAAAAGUGCUCGAGAG_CCAUAAACCUCUGCAAAAGACUCAACUUUUGUUAGGACUAAGCAGCUUAUAUCAG ....((((((((((.(((((((...........))).............((...((.((((......)))).))..)).)))).))))))))))... (-14.94 = -15.55 + 0.61)

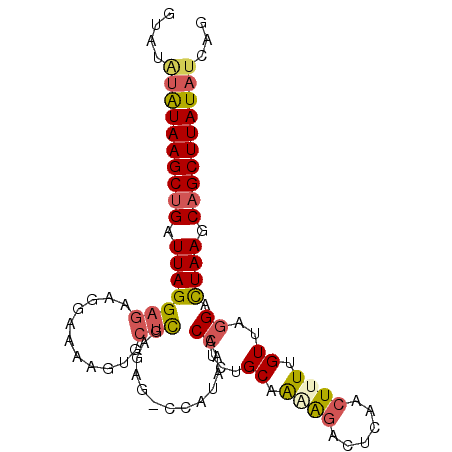
| Location | 5,497,343 – 5,497,439 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -11.60 |
| Energy contribution | -13.41 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5497343 96 - 22407834 CUGAUAUAAGCUGCUUAGUCCUAACAAAAGUUGAGUCUUUGGCAAAGGUUUAUGG-CUCUCGAGCACUUUUCCUUCUCCUAAUCAGCUUAUACAUAU .((.(((((((((.((((.......((((((.(((((...(((....)))...))-)))......)))))).......)))).)))))))))))... ( -22.94) >DroPse_CAF1 136912 81 - 1 CUGCUAUAAGCAGCUUAAACUAAACAAAAGUUGGGGCUGUUGCAGAGGUCUGAGGUCUCUGAAG-------------CCUAAUCAGCUUACGCA--- ((((.....))))..............(((((((((((....(((((..(...)..))))).))-------------)))...)))))).....--- ( -25.30) >DroSec_CAF1 121128 96 - 1 CUGAUAUAAGCUGCUUAGUCCUAACAAAAGUUGAGUCUUUGGCAGAGGUUUAUGG-CUUUUGAGCACUUUUCCUUCUCCUAAUCAGCUUAUAUAUAC ...((((((((((.((((....(((....)))(((.....((.(((((((((...-....))))).)))).))..))))))).)))))))))).... ( -24.20) >DroSim_CAF1 127926 96 - 1 CUGAUAUAAGCUGCUUAGUCCUAACAAAAGUUAAGUCUUUGGCAAAGGUUUAUGG-CUCUCGAGCACUUUUCCUUCUCCUAAUCAGCUUAUAUAUAC ...((((((((((.((((.(.((((....)))).).)...((..((((......(-((....)))......))))..))))).)))))))))).... ( -23.70) >DroEre_CAF1 118838 93 - 1 CUGAUAUAAGCUGCUUAGUCCUAACAAAA---GAGUCCGCUGCACAGGUUUAUGG-CUCUCGAGCACUUUUUCCUCUCCUAAUCAGCUUAUAUAUAC ...((((((((((.((((.......((((---(.(..((..((.((......)))-)...))..).))))).......)))).)))))))))).... ( -23.54) >DroYak_CAF1 125606 96 - 1 CUGAUAUAAGCUGCUUAGUCCUAACAAAAGUUGAGUCUCUUGCACAGGUUUAUGG-CUCUCGAGCACUUUUUCUUCUCCUAAUCAGCUUAUAUAUAC ...((((((((((.((((.......((((((.(((((....((....))....))-)))......)))))).......)))).)))))))))).... ( -23.14) >consensus CUGAUAUAAGCUGCUUAGUCCUAACAAAAGUUGAGUCUUUGGCAAAGGUUUAUGG_CUCUCGAGCACUUUUCCUUCUCCUAAUCAGCUUAUAUAUAC ...((((((((((.((((.......((((((.(((.((...((....))....)).)))......)))))).......)))).)))))))))).... (-11.60 = -13.41 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:42 2006