| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,494,408 – 5,494,517 |
| Length | 109 |
| Max. P | 0.999870 |

| Location | 5,494,408 – 5,494,517 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.32 |
| SVM RNA-class probability | 0.999870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5494408 109 + 22407834 UGAAAUUGCCAACAUGCCAGAGAUUACCAACUGUCAGUUGAAUGUCAUGCCAAUGGAACAUGCCGUCAACUGAAGUGUUGAACUAAUUUAACAGCUGUUGAAA-AAAAAU-- .........(((((.((...((((((.((((..((((((((.((.((((((...))..)))).))))))))))...))))...))))))....)))))))...-......-- ( -26.80) >DroVir_CAF1 166228 111 + 1 AGAAAUUGCCAACAUGCCAGAGAUUACCAACUGUCAGUUGAAUGUCAUGCCAAUGGAACAUGCCGUCAACUGAAGUGUUGAACUAAUUUAGCAGCUGUUGAAA-AAAGCCAA .........((((((((...((((((.((((..((((((((.((.((((((...))..)))).))))))))))...))))...)))))).)))..)))))...-........ ( -26.80) >DroGri_CAF1 176341 111 + 1 AGAAAUUGCCAACAUGCCAGAGAUUACCAACUGUCAGUUGAAUGUCAUGCCGUUGGAACAUGCCGUCAACUGAAGUGUUGAACUAAUUUAGCAGCUGUUGAAA-AAAGCCAA .........((((((((...((((((.((((..((((((((.((.((((((...))..)))).))))))))))...))))...)))))).)))..)))))...-........ ( -26.80) >DroSim_CAF1 125025 109 + 1 UGAAAUUGCCAACAUGCCAGAGAUUACCAACUGUCAGUUGAAUGUCAUGCCAAUGGAACAUGCCGUCAACUGAAGUGUUGAACUAAUUUAACAGCUGUUGAAA-AAAAAU-- .........(((((.((...((((((.((((..((((((((.((.((((((...))..)))).))))))))))...))))...))))))....)))))))...-......-- ( -26.80) >DroWil_CAF1 168185 109 + 1 AGAAAUUGCCAACAUGCCAGAGAUUACCAACUGUCAGUUGAAUGUCAUGCCAAUGGAACAUGCCGUCAACUGAAGUGUUGAACUAAUUUAACAGCUGUUGAAA-AAAGAA-- .........(((((.((...((((((.((((..((((((((.((.((((((...))..)))).))))))))))...))))...))))))....)))))))...-......-- ( -26.80) >DroMoj_CAF1 184602 112 + 1 AGAAAUUGCCAACAUGCCAGAGAUUACCAACUGUCAGUUGAAUGUCAUGCCAAUGGAACAUGCCGUCAACUGAAGUGUUGAACUAAUUUAGCAGCUGUUGAAAAAAAGCCAA .........((((((((...((((((.((((..((((((((.((.((((((...))..)))).))))))))))...))))...)))))).)))..)))))............ ( -26.80) >consensus AGAAAUUGCCAACAUGCCAGAGAUUACCAACUGUCAGUUGAAUGUCAUGCCAAUGGAACAUGCCGUCAACUGAAGUGUUGAACUAAUUUAACAGCUGUUGAAA_AAAGAC__ .........(((((.((...((((((.((((..((((((((.((.((((((...))..)))).))))))))))...))))...))))))....)))))))............ (-26.80 = -26.80 + 0.00)


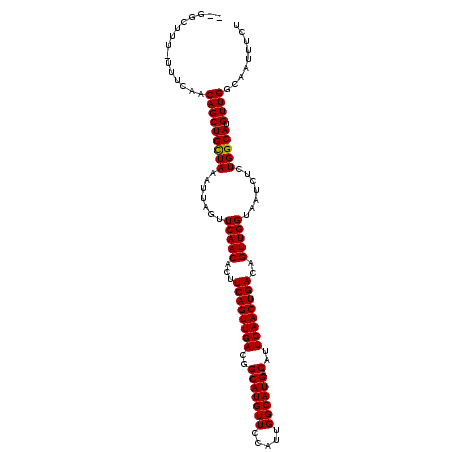
| Location | 5,494,408 – 5,494,517 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.61 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.18 |
| SVM RNA-class probability | 0.999827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5494408 109 - 22407834 --AUUUUU-UUUCAACAGCUGUUAAAUUAGUUCAACACUUCAGUUGACGGCAUGUUCCAUUGGCAUGACAUUCAACUGACAGUUGGUAAUCUCUGGCAUGUUGGCAAUUUCA --......-..(((((((((.....((((..(((((...((((((((..(((((((.....)))))).)..))))))))..)))))))))....))).))))))........ ( -26.50) >DroVir_CAF1 166228 111 - 1 UUGGCUUU-UUUCAACAGCUGCUAAAUUAGUUCAACACUUCAGUUGACGGCAUGUUCCAUUGGCAUGACAUUCAACUGACAGUUGGUAAUCUCUGGCAUGUUGGCAAUUUCU ........-......(((((((((.......(((((...((((((((..(((((((.....)))))).)..))))))))..))))).......))))).))))......... ( -28.84) >DroGri_CAF1 176341 111 - 1 UUGGCUUU-UUUCAACAGCUGCUAAAUUAGUUCAACACUUCAGUUGACGGCAUGUUCCAACGGCAUGACAUUCAACUGACAGUUGGUAAUCUCUGGCAUGUUGGCAAUUUCU ........-......(((((((((.......(((((...((((((((..(((((((.....)))))).)..))))))))..))))).......))))).))))......... ( -28.94) >DroSim_CAF1 125025 109 - 1 --AUUUUU-UUUCAACAGCUGUUAAAUUAGUUCAACACUUCAGUUGACGGCAUGUUCCAUUGGCAUGACAUUCAACUGACAGUUGGUAAUCUCUGGCAUGUUGGCAAUUUCA --......-..(((((((((.....((((..(((((...((((((((..(((((((.....)))))).)..))))))))..)))))))))....))).))))))........ ( -26.50) >DroWil_CAF1 168185 109 - 1 --UUCUUU-UUUCAACAGCUGUUAAAUUAGUUCAACACUUCAGUUGACGGCAUGUUCCAUUGGCAUGACAUUCAACUGACAGUUGGUAAUCUCUGGCAUGUUGGCAAUUUCU --......-..(((((((((.....((((..(((((...((((((((..(((((((.....)))))).)..))))))))..)))))))))....))).))))))........ ( -26.50) >DroMoj_CAF1 184602 112 - 1 UUGGCUUUUUUUCAACAGCUGCUAAAUUAGUUCAACACUUCAGUUGACGGCAUGUUCCAUUGGCAUGACAUUCAACUGACAGUUGGUAAUCUCUGGCAUGUUGGCAAUUUCU ...............(((((((((.......(((((...((((((((..(((((((.....)))))).)..))))))))..))))).......))))).))))......... ( -28.84) >consensus __GGCUUU_UUUCAACAGCUGCUAAAUUAGUUCAACACUUCAGUUGACGGCAUGUUCCAUUGGCAUGACAUUCAACUGACAGUUGGUAAUCUCUGGCAUGUUGGCAAUUUCU ...............(((((((((.......(((((...((((((((..(((((((.....)))))).)..))))))))..))))).......))))).))))......... (-27.86 = -27.61 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:38 2006