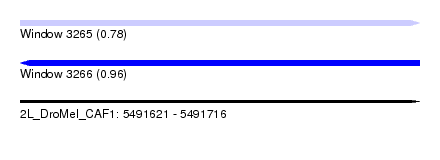
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,491,621 – 5,491,716 |
| Length | 95 |
| Max. P | 0.960614 |

| Location | 5,491,621 – 5,491,716 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -11.54 |
| Energy contribution | -11.43 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
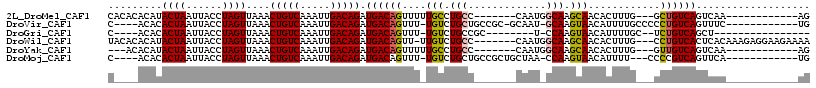
>2L_DroMel_CAF1 5491621 95 + 22407834 CACACACAUACUAAUUACCUAGUUAAACUGUCAAAUUGACAGAUGACAGUUUUUGCCUGCC-------CAAUGGCAAGCAACACUUUG---GCUGUCAGUCAA------------AG .........((((......))))....(((((.....))))).((((((((.((((.((((-------....)))).))))......)---))))))).....------------.. ( -25.50) >DroVir_CAF1 162687 98 + 1 C----ACACACUAAUUACCUAGUUAAACUGUCAAAUUGACAGAUGACAGUUU-UGUCUGCUGCCGC-GCAAU-GCAAGUAACAUUUUGCCCCCUGUCAGUUUC------------UG .----....((((......))))....(((((.....))))).((((((...-....(((......-)))..-(((((......)))))...)))))).....------------.. ( -20.40) >DroGri_CAF1 172514 85 + 1 C----ACACACUAAUUACCUAGUUAAACUGUCAAAUUGACAGAUGACAGUUU-UGUCUGCCGC--------U-CCAAGUAACAUUUUGC--UCUGUCAGCU---------------- .----....((((......))))....(((((.....))))).((((((...-(((.(((...--------.-....))))))......--.))))))...---------------- ( -15.20) >DroWil_CAF1 163018 106 + 1 UACACACAUACUAAUUACCUAGUUAAACUGUCAAAUUGACAGAUGACAGUU-UUGUCUGCC-------CAAUGGCAAGCAACACUUUG---CCUGUCACUCACAAAGAGGAAGAAAA .(((.....((((......)))).....))).....((((((..((.(((.-(((..((((-------....))))..))).))))).---.))))))(((.....)))........ ( -23.80) >DroYak_CAF1 119806 92 + 1 ---ACACAUACUAAUUACCUAGUUAAACUGUCAAAUUGACAGAUGACAGUUUUUGCCUGCC-------CAAUGGCAAGCAACACUUUG---GUUGUCAGUCAA------------AG ---......(((.((.(((.(((.(((((((((..........)))))))))((((.((((-------....)))).)))).)))..)---)).)).)))...------------.. ( -23.20) >DroMoj_CAF1 180172 96 + 1 C----ACACACUAAUUACCUAGUUAAACUGUCAAAUUGACAGAUGACAGUUU-UGUCUGCUGCCGCUGCUAA-CCAAGUAACAUUUU---CCCCGUCAGUUCA------------UG .----....((((......))))....(((((.....))))).(((((((..-.....)))...(.((((..-...)))).).....---....)))).....------------.. ( -13.30) >consensus C____ACACACUAAUUACCUAGUUAAACUGUCAAAUUGACAGAUGACAGUUU_UGUCUGCC_______CAAU_GCAAGCAACACUUUG___CCUGUCAGUCAA____________AG .........((((......))))....(((((.....))))).((((((....(((.(((.............))).)))............))))))................... (-11.54 = -11.43 + -0.11)

| Location | 5,491,621 – 5,491,716 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
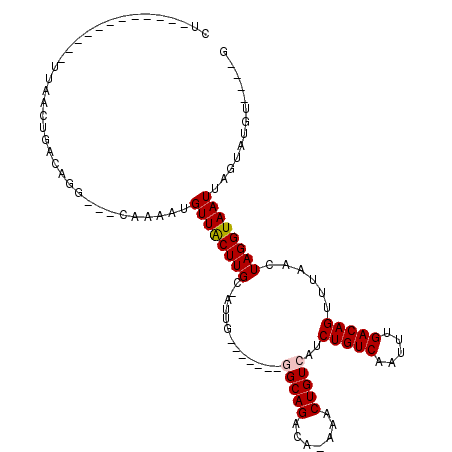
>2L_DroMel_CAF1 5491621 95 - 22407834 CU------------UUGACUGACAGC---CAAAGUGUUGCUUGCCAUUG-------GGCAGGCAAAAACUGUCAUCUGUCAAUUUGACAGUUUAACUAGGUAAUUAGUAUGUGUGUG ..------------...(((((..((---(......(((((((((....-------)))))))))(((((((((..........))))))))).....)))..)))))......... ( -32.00) >DroVir_CAF1 162687 98 - 1 CA------------GAAACUGACAGGGGGCAAAAUGUUACUUGC-AUUGC-GCGGCAGCAGACA-AAACUGUCAUCUGUCAAUUUGACAGUUUAACUAGGUAAUUAGUGUGU----G ..------------.((((((.(((((.(((..((((.....))-)))))-.)(((((..((((-....))))..)))))..)))).)))))).(((((....)))))....----. ( -25.20) >DroGri_CAF1 172514 85 - 1 ----------------AGCUGACAGA--GCAAAAUGUUACUUGG-A--------GCGGCAGACA-AAACUGUCAUCUGUCAAUUUGACAGUUUAACUAGGUAAUUAGUGUGU----G ----------------..........--(((....(((((((((-.--------..(((((...-...)))))..(((((.....))))).....)))))))))...)))..----. ( -23.00) >DroWil_CAF1 163018 106 - 1 UUUUCUUCCUCUUUGUGAGUGACAGG---CAAAGUGUUGCUUGCCAUUG-------GGCAGACAA-AACUGUCAUCUGUCAAUUUGACAGUUUAACUAGGUAAUUAGUAUGUGUGUA .......((.....(..((..(((..---.....)))..))..)....)-------)(((.((((-((((((((..........))))))))).(((((....))))).))).))). ( -28.90) >DroYak_CAF1 119806 92 - 1 CU------------UUGACUGACAAC---CAAAGUGUUGCUUGCCAUUG-------GGCAGGCAAAAACUGUCAUCUGUCAAUUUGACAGUUUAACUAGGUAAUUAGUAUGUGU--- ..------------...(((((..((---(......(((((((((....-------)))))))))(((((((((..........))))))))).....)))..)))))......--- ( -31.70) >DroMoj_CAF1 180172 96 - 1 CA------------UGAACUGACGGGG---AAAAUGUUACUUGG-UUAGCAGCGGCAGCAGACA-AAACUGUCAUCUGUCAAUUUGACAGUUUAACUAGGUAAUUAGUGUGU----G ((------------((.((((......---.....(((((((((-((((..((....)).((((-....))))..(((((.....))))).))))))))))))))))).)))----) ( -23.70) >consensus CU____________UUAACUGACAGG___CAAAAUGUUACUUGC_AUUG_______GGCAGACA_AAACUGUCAUCUGUCAAUUUGACAGUUUAACUAGGUAAUUAGUAUGU____G ...................................((((((((.............(((((.......)))))..(((((.....)))))......))))))))............. (-14.87 = -14.95 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:31 2006