
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,483,071 – 5,483,176 |
| Length | 105 |
| Max. P | 0.746102 |

| Location | 5,483,071 – 5,483,176 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5483071 105 - 22407834 AUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUG---GCAGGAUACACUGCAA------CUGCAAC----ACCA-- .....((((((((....(((((....(((((((((((........)))........)).))))))....)))))))))---((((......))))..------))))...----....-- ( -36.50) >DroVir_CAF1 149623 100 - 1 GCU-GGCAGCGUAAAUUGAGUUUAUUGCGCCGCC---GUAUUGCAGGCAAAUUACAGGUCGGCGCAUUUAACUUUGUG---CCAGGAUAUG---AAA------GGG-GGC--AGAGACA- .((-((((.((......(((((...(((((((((---((((((....)))..))).)).)))))))...)))))))))---))))......---...------...-...--.......- ( -31.40) >DroGri_CAF1 159943 107 - 1 GCCGGGCAGCGUAAAUUGAGUUUAUUGCGCCGCC---GUAUUGCUGGCAAAUUACAGGUCGGCGCAUUUAACUUUGUGCUGCCAGGAUAUG---AAA------GGG-GGCCCAGAAACAG .((.(((((((((....(((((...(((((((((---((((((....)))..))).)).)))))))...)))))))))))))).)).....---...------((.-...))........ ( -40.00) >DroEre_CAF1 103465 111 - 1 AUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUG---GCAGGAUACACUGCAACUGCAACUGCAAC----ACUA-- .....((((((((....(((((....(((((((((((........)))........)).))))))....)))))))))---((((......))))..)))).........----....-- ( -36.50) >DroWil_CAF1 149331 101 - 1 AUCGGUCAGAGCAAAUUGAGUUUAUUGCGACGCC---GUAUUACAGGCAAAUUACAGGUCGGCACAUUUUACUUUGUG---CCAGGAUAGG----AA------GGGGAGCG-AGAGAA-- .(((.((.((.(.......(((......)))(((---........)))........).))((((((........))))---))........----..------...)).))-).....-- ( -24.50) >DroPer_CAF1 121928 105 - 1 AUCGAGCAGCACAAAUUGAGUUUACUGCGCCGACGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUG---GCAGGAUACCCUGUAA------GUGCAAC----UGCA-- .....((((((((....(((((....(((((((((((........))).........))))))))....)))))))))---(((((....)))))..------......)----))).-- ( -38.30) >consensus AUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCC___GUAUUGCUGGCAAAUUACAGGUCGGCGCAUUUAACUUUGUG___CCAGGAUACG___AAA______GGGCAAC____AGAA__ .((......((((....(((((....((((((((......(((....)))......)).))))))....)))))))))......)).................................. (-15.53 = -15.70 + 0.17)

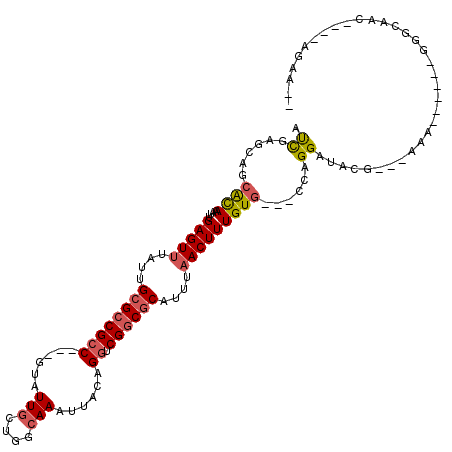
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:29 2006