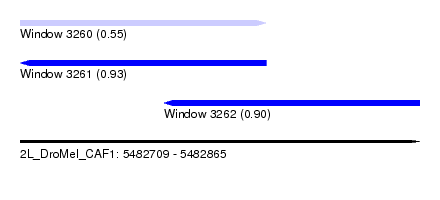
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,482,709 – 5,482,865 |
| Length | 156 |
| Max. P | 0.928070 |

| Location | 5,482,709 – 5,482,805 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
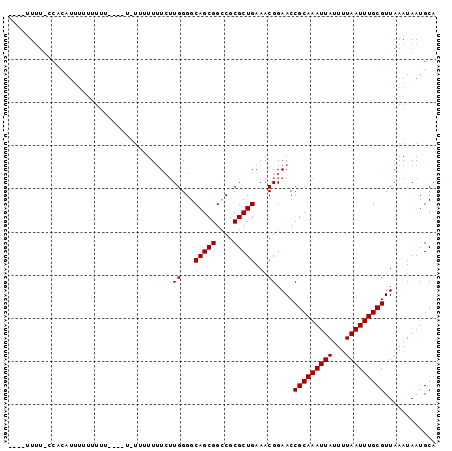
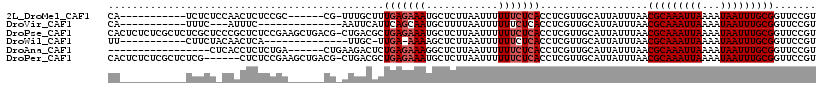
>2L_DroMel_CAF1 5482709 96 + 22407834 UUUUUUUUGCCGCAUUCUUUUUUUUUAUU---UUUUCAUGGGGCAGCGGUCGCGCUGAAACGGAACCGCAAAUUAUUUUAAUUUGCGUUAAAUAAUGCA ...........(((((.(((.........---..(((.((...(((((....)))))...))))).(((((((((...)))))))))..))).))))). ( -21.70) >DroSec_CAF1 101885 89 + 1 ----UUUUGCCGCAUUUUUUUUU------UUAUUUUCUUGGGGCAGCGGCCGCGCUGAAACGGAACCGCAAAUUAUUUUAAUUUGCGUUAAAUAAUGCA ----.......(((((.(((...------.....((((.(...(((((....)))))...))))).(((((((((...)))))))))..))).))))). ( -21.90) >DroEre_CAF1 103131 84 + 1 -----UUU-CCAUAUAUAC--------U-UUUUUUUCUUGGGGCAGCGGCCGCGCUGAAACGGAACCGCAAAUUAUUUUAAUUUGCGUUAAAUAAUGCA -----.((-((.......(--------(-((........))))(((((....)))))....)))).(((((((((...)))))))))............ ( -18.20) >DroYak_CAF1 110512 93 + 1 -----UUUACCAUAUAUCUUUCUUUCUU-UUUUUUUCUUGGGGCAGCGGCCGCGCUGAAACGGAACCGCAAAUUAUUUUAAUUUGCGUUAAAUAAUGCA -----......................(-((..((((.((.(((....))).))..))))..))).(((((((((...)))))))))............ ( -17.10) >DroAna_CAF1 109526 81 + 1 ----UUC--------UUUUCUUA------UUGUUGUGUUGGGGCAGCGGCCGCGCUGAAACGGAACCGCAAAUUAUUUUAAUUUGCGUUAAAUAAUGCA ----...--------.....(((------(((((.((((....(((((....))))).)))).)))(((((((((...)))))))))...))))).... ( -20.80) >consensus ____UUUU_CCACAUUUUUUUUU____U_UUUUUUUCUUGGGGCAGCGGCCGCGCUGAAACGGAACCGCAAAUUAUUUUAAUUUGCGUUAAAUAAUGCA ......................................((...(((((....)))))...))....(((((((((...)))))))))............ (-15.62 = -15.62 + 0.00)

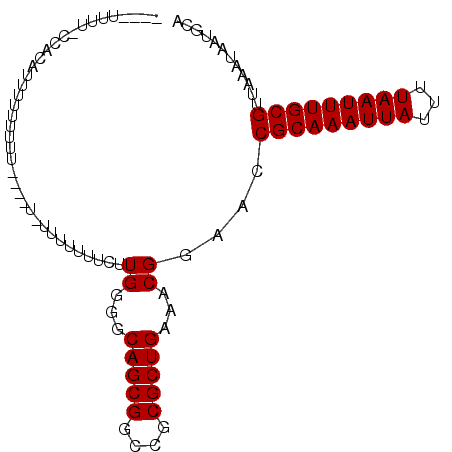
| Location | 5,482,709 – 5,482,805 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -18.36 |
| Consensus MFE | -16.10 |
| Energy contribution | -15.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
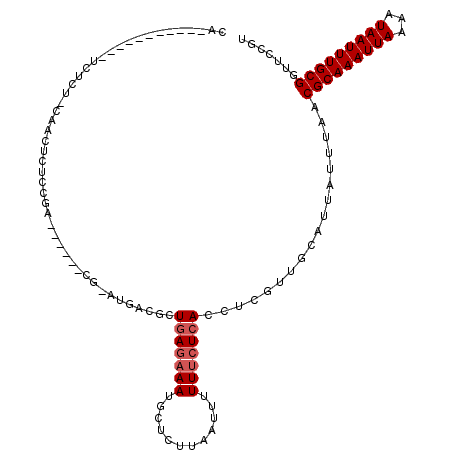
>2L_DroMel_CAF1 5482709 96 - 22407834 UGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGUUUCAGCGCGACCGCUGCCCCAUGAAAA---AAUAAAAAAAAAGAAUGCGGCAAAAAAAA .((..........((((((((...))))))))((((.(((....))).))))))((((.(((.....---..............))).))))....... ( -22.11) >DroSec_CAF1 101885 89 - 1 UGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGUUUCAGCGCGGCCGCUGCCCCAAGAAAAUAA------AAAAAAAAAUGCGGCAAAA---- .((..........((((((((...))))))))(((..(((....)))..)))))((((.((.........------.........)).))))...---- ( -19.97) >DroEre_CAF1 103131 84 - 1 UGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGUUUCAGCGCGGCCGCUGCCCCAAGAAAAAAA-A--------GUAUAUAUGG-AAA----- ............(((((((((...))))))))).((((((..(((((....)))))..............-.--------......))))-)).----- ( -17.20) >DroYak_CAF1 110512 93 - 1 UGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGUUUCAGCGCGGCCGCUGCCCCAAGAAAAAAA-AAGAAAGAAAGAUAUAUGGUAAA----- (((..(((((..(((((((((...))))))))).(((.....(((((....)))))......))).....-.........)))))....)))..----- ( -16.50) >DroAna_CAF1 109526 81 - 1 UGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGUUUCAGCGCGGCCGCUGCCCCAACACAACAA------UAAGAAAA--------GAA---- .((..........((((((((...))))))))(((..(((....)))..)))))................------........--------...---- ( -16.00) >consensus UGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGUUUCAGCGCGGCCGCUGCCCCAAGAAAAAAA_A____AAAAAAAAAUACGG_AAAA____ .((..........((((((((...))))))))(((..(((....)))..)))))............................................. (-16.10 = -15.94 + -0.16)

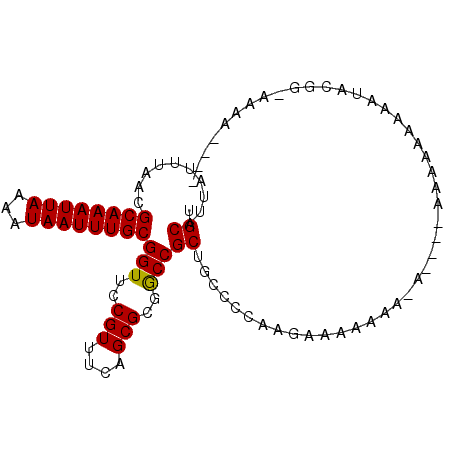
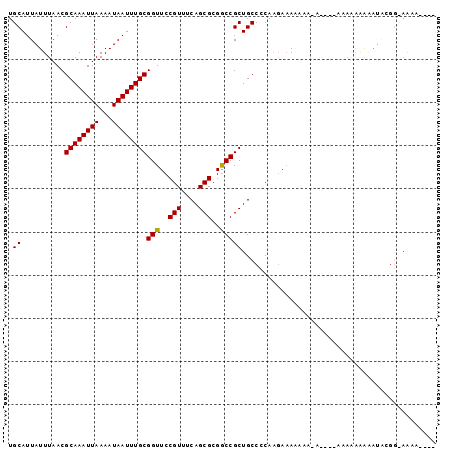
| Location | 5,482,765 – 5,482,865 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -17.81 |
| Consensus MFE | -11.05 |
| Energy contribution | -11.55 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
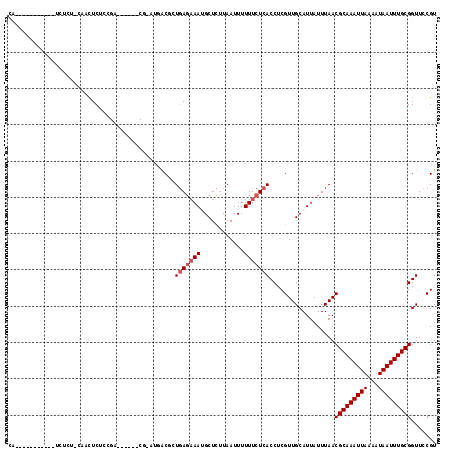
>2L_DroMel_CAF1 5482765 100 - 22407834 CA-----------UCUCUCCAACUCUCCGC------CG-UUUGCUUUGAGAAAUGCUCUUAAUUUUUUCUCACCUCGUUGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGU ..-----------......((((.....((------..-...))..(((((((............)))))))....))))..........(((((((((...)))))))))....... ( -16.90) >DroVir_CAF1 148926 90 - 1 CA-----------UUUC---AUUUC--------------AAUUCAUUCAGCAAUGCUUUUAAUUUUUUCUCACCUCGUUGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGU ..-----------....---.....--------------..........((((((....................)))))).........(((((((((...)))))))))....... ( -12.95) >DroPse_CAF1 120941 117 - 1 CACUCUCUCGCUCUCGCUCCCGCUCUCCGAAGCUGACG-CUGACGCUGAGAAAUGCUCUUAAUUUUUUCUCACCUCGUUGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGU .........(((.(((...........)))))).((((-(.((((.(((((((............)))))))...)))))).........(((((((((...)))))))))))).... ( -22.70) >DroWil_CAF1 149121 91 - 1 UU-----------CUUCUACAACUCA--------------UUGC-UUGA-AAAAGCUCUUAAUUUUUUCUCACCUCGUUGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGU ..-----------.............--------------..((-((..-..))))...............(((..((((.......))))((((((((...)))))))))))..... ( -14.00) >DroAna_CAF1 109567 95 - 1 -----------------CUCACCUCUCUGA------CUGAAGACUCUGAGAAAGGCUCUUAAUUUUUUCUCACCUCGUUGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGU -----------------............(------(.(((((...(((((((((........)))))))))..))..............(((((((((...))))))))).))).)) ( -16.60) >DroPer_CAF1 121686 111 - 1 CACUCUCUCGCUCUCG------CUCUCCGAAGCUGACG-CUGACGCUGAGAAAUGCUCUUAAUUUUUUCUCACCUCGUUGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGU .........(((.(((------.....)))))).((((-(.((((.(((((((............)))))))...)))))).........(((((((((...)))))))))))).... ( -23.70) >consensus CA___________UCUCU_CAACUCUCCGA______CG_AUGACGCUGAGAAAUGCUCUUAAUUUUUUCUCACCUCGUUGCAUUAUUUAACGCAAAUUAAAAUAAUUUGCGGUUCCGU ..............................................(((((((............)))))))..................(((((((((...)))))))))....... (-11.05 = -11.55 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:28 2006