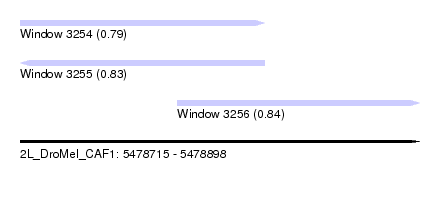
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,478,715 – 5,478,898 |
| Length | 183 |
| Max. P | 0.835717 |

| Location | 5,478,715 – 5,478,827 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5478715 112 + 22407834 UCGGUGGCGGU---GGUAUCUGAAA--UAUACGAG-AGUCCCCGGUUGGCAGUUACCGGUUACCGGUUGUGGGUCCAAGGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAA ((((.(((..(---.((((......--.)))).).-.))).))))((((((((((((((...))))).((((((.........))))))(((((....))))).....))))))))). ( -36.70) >DroPse_CAF1 116694 98 + 1 ------------------CAUGGUAUG-GUCCGUUUGGUCCCCGGUCGCCAGUUACCGGUUACCGGG-GUCGGUCCCAGGUAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAA ------------------..(((((.(-(.(((......(((((((.(((.......))).))))))-).))).))..((.....))(((((((....)))))))......))))).. ( -37.00) >DroEre_CAF1 99081 108 + 1 UCGGUGG------CGGUGUCUGAAA--UAUACGAG-AGUCCCCGGUUGGCAGUUACCGGUUACCGGUUGUGGGUCCA-GGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAA ....(((------(((((((((...--..(((((.-.((..(((((........)))))..))...)))))....))-)))......(((((((....)))))))....))))))).. ( -35.70) >DroYak_CAF1 106506 115 + 1 UCGGUGGCGGUGGCGGUGUGUGAAA--UAUACGAG-AGUCCCCGGUUGGCAGUUACCGGUUACCGGUUGUGGGUCCAAGGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAA .(((((((((.(((..((((((...--))))))..-.)))))((((........)))))))))))(((((((((.........))))(((((((....))))))).)))))....... ( -37.80) >DroAna_CAF1 105884 115 + 1 UCGGUGGCACU---GUAUUCCGGAAUAUAUACGAGAAGUCCCCGGUUGGCAGUUACCGGUUGCCGGUUGUGGGUCCGAGGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAA ((((.((.(((---....((((.........)).)))))))))))(((((((((...((.((((((........))..))))...))(((((((....)))))))...))))))))). ( -37.10) >DroPer_CAF1 117406 98 + 1 ------------------CAUGGUAUG-GUCCGUUUGGUCCCCGGUCGCCAGUUACCGGUUACCGGG-GUCGGUCCCAGGUAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAA ------------------..(((((.(-(.(((......(((((((.(((.......))).))))))-).))).))..((.....))(((((((....)))))))......))))).. ( -37.00) >consensus UCGGUGG_______GGU_UCUGAAA__UAUACGAG_AGUCCCCGGUUGGCAGUUACCGGUUACCGGUUGUGGGUCCAAGGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAA .....................................((..(((((........)))))..))(((((((........((.....))(((((((....))))))).)))))))..... (-22.95 = -23.48 + 0.53)

| Location | 5,478,715 – 5,478,827 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -32.41 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5478715 112 - 22407834 UUUAGCAGUUGUUAUGAGUGUUAACACUCAUGGAUAUGCCUUGGACCCACAACCGGUAACCGGUAACUGCCAACCGGGGACU-CUCGUAUA--UUUCAGAUACC---ACCGCCACCGA ....((((((.(((((((((....)))))))))..................(((((...)))))))))))....(((((...-...((((.--......)))).---....)).))). ( -31.00) >DroPse_CAF1 116694 98 - 1 UUUAGCAGUUGUUAUGAGUGUUAACACUCAUGGAUAUACCUGGGACCGAC-CCCGGUAACCGGUAACUGGCGACCGGGGACCAAACGGAC-CAUACCAUG------------------ .....(((.(((((((((((....))))))).))))...)))((.(((.(-((((((..((((...))))..)))))))......))).)-)........------------------ ( -35.50) >DroEre_CAF1 99081 108 - 1 UUUAGCAGUUGUUAUGAGUGUUAACACUCAUGGAUAUGCC-UGGACCCACAACCGGUAACCGGUAACUGCCAACCGGGGACU-CUCGUAUA--UUUCAGACACCG------CCACCGA ....(((....(((((((((....)))))))))...)))(-((((.........(((..(((((........)))))..)))-........--.)))))......------....... ( -29.57) >DroYak_CAF1 106506 115 - 1 UUUAGCAGUUGUUAUGAGUGUUAACACUCAUGGAUAUGCCUUGGACCCACAACCGGUAACCGGUAACUGCCAACCGGGGACU-CUCGUAUA--UUUCACACACCGCCACCGCCACCGA ....((.(((((((((((((....))))))))(((((((...((........))(((..(((((........)))))..)))-...)))))--))..))).)).))............ ( -29.20) >DroAna_CAF1 105884 115 - 1 UUUAGCAGUUGUUAUGAGUGUUAACACUCAUGGAUAUGCCUCGGACCCACAACCGGCAACCGGUAACUGCCAACCGGGGACUUCUCGUAUAUAUUCCGGAAUAC---AGUGCCACCGA ....(((....(((((((((....)))))))))...))).((((.......(((((...))))).((((....((((((((.....)).....))))))....)---)))....)))) ( -33.70) >DroPer_CAF1 117406 98 - 1 UUUAGCAGUUGUUAUGAGUGUUAACACUCAUGGAUAUACCUGGGACCGAC-CCCGGUAACCGGUAACUGGCGACCGGGGACCAAACGGAC-CAUACCAUG------------------ .....(((.(((((((((((....))))))).))))...)))((.(((.(-((((((..((((...))))..)))))))......))).)-)........------------------ ( -35.50) >consensus UUUAGCAGUUGUUAUGAGUGUUAACACUCAUGGAUAUGCCUUGGACCCACAACCGGUAACCGGUAACUGCCAACCGGGGACU_CUCGUAUA__UUCCAGA_ACC_______CCACCGA ....(((....(((((((((....)))))))))...)))...((........))(((..(((((........)))))..))).................................... (-24.17 = -24.45 + 0.28)



| Location | 5,478,787 – 5,478,898 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.94 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -23.18 |
| Energy contribution | -23.35 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5478787 111 + 22407834 GGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAAGCCCAUUUUCGCAUGUUGGCAGCUUAUAAAAGUGCAGCACUCGGCCGCAUGGCCAUAUACC---------ACAUAUAUAG ((.((((....(((((((..((((........(((((((.((........))...)))))))........)))).)))))))((((....)))))))).))---------.......... ( -31.39) >DroPse_CAF1 116752 100 + 1 GGUAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAAGCCCAUUUUCGCAUGUUGGCAGCUUAUAAAACUGCAGCACUCGGCCAUAGAGCCAUA--------------------UAG ((.....))..(((((((...........(((((((.((((....)))).))).))))((((.........)))))))))))(((......)))...--------------------... ( -25.30) >DroEre_CAF1 99149 109 + 1 GGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAAGCCCAUUUUCGCAUGUUGGCAGCUUAUAAAAGUGCAGCGCUCGGCGGUAUGGCCAUAUACC---------ACA--UACAG ((((((((..((((((((..((((........(((((((.((........))...)))))))........)))).))))))))..))))).))).......---------...--..... ( -30.49) >DroWil_CAF1 144432 100 + 1 GGUAUAUCCAUGAGUGUUAACAUUCAUAACAACUGCUAAAGCCCAUUUUCGCAUGUUGGCAGCUUAUAAAAAUGCAGCACUCGACCAUAGCACCAUA--------------------UAG (((.(((...((((((((..((((........(((((((.((........))...)))))))........)))).))))))))...)))..)))...--------------------... ( -22.69) >DroAna_CAF1 105959 120 + 1 GGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAAGCCCAUUUUCGCAUGUUGGCAGCUUAUAAAAAUGCAGCACUCGGCCGUGGAGCCAUACAUCCCAUUAGCAACAUAUAUAG .........(((((((....)))))))......((((((.((........))((((((((.((..........))..(((......)))..)))).))))....)))))).......... ( -26.70) >DroPer_CAF1 117464 100 + 1 GGUAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAAGCCCAUUUUCGCAUGUUGGCAGCUUAUAAAACUGCAGCACUCGGCCAUAGAGCCAUA--------------------UAG ((.....))..(((((((...........(((((((.((((....)))).))).))))((((.........)))))))))))(((......)))...--------------------... ( -25.30) >consensus GGCAUAUCCAUGAGUGUUAACACUCAUAACAACUGCUAAAGCCCAUUUUCGCAUGUUGGCAGCUUAUAAAAAUGCAGCACUCGGCCAUAGAGCCAUA____________________UAG .((((....(((((((....))))))).....(((((((.((........))...))))))).........)))).......(((......))).......................... (-23.18 = -23.35 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:22 2006