
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,473,242 – 5,473,332 |
| Length | 90 |
| Max. P | 0.609847 |

| Location | 5,473,242 – 5,473,332 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.26 |
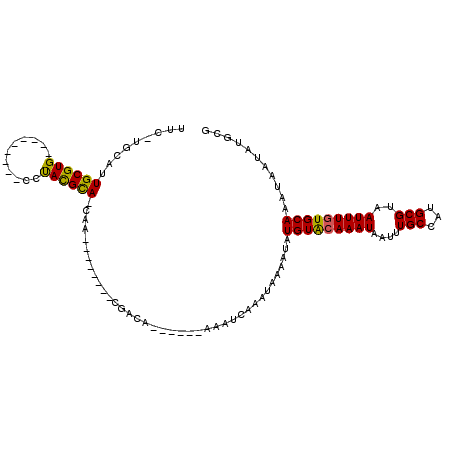
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -13.39 |
| Energy contribution | -12.92 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609847 |
| Prediction | RNA |
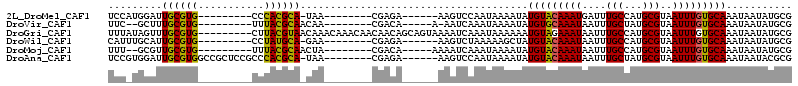
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5473242 90 + 22407834 UCCAUGGAUUGCGUG---------CCCACGCA-UAA--------CGAGA------AAGUCCAAUAAAAUAUGUACAAAUGAUUUGCCAUGCGUAAUUUGUGCAAAUAAUAUGCG ....((((((((((.---------...)))))-...--------.....------..)))))........(((((((((....(((...)))..)))))))))........... ( -19.50) >DroVir_CAF1 136184 89 + 1 UUC--GCUUUGCGUG---------UUUACGCAACAA--------CGACA-----A-AAUCAAAUAAAAUAUGUGCAAAUAAUUUGCUAUGCGUAAUUUGUGCAAAUAAUAUGCG .((--(..((((((.---------...))))))...--------)))..-----.-.................(((.((.((((((...(((.....))))))))).)).))). ( -16.60) >DroGri_CAF1 143509 105 + 1 UUUAUAGUUUGCGUG---------CUUACGUAACAAACAAACAACAACAGCAGUAAAAUCAAAUAAAAAAUGUAGAAAUAAUUUGCCAUGCGUAAUUUGUGCAAAUAAUAUGCG ..(((.(((((((..---------.(((((((..............((....)).................(((((.....)))))..)))))))....))))))).))).... ( -12.80) >DroWil_CAF1 138975 90 + 1 CAUUUGCAUUGCGUG---------CCUAUGCA-GAA--------CGAGA------AAGUCUAAAAAGCUAUGUACAAAUAAUUUGCCAUGCGUAAUUUGUGCAAAUAAUAUGCG .....(((((((((.---------...)))))-)..--------.....------...............(((((((((....(((...)))..))))))))).......))). ( -15.90) >DroMoj_CAF1 150674 90 + 1 UUU--GCGUUGCGUG---------UUUACGCAACUA--------CGACA-----AAAAUCAAAUAAAAUAUGUACAAAUAAUUUGCCAUGCGUAAUUUGUGCAAAUAAUAUGCG ...--(((((((((.---------...)))))))..--------.....-----................(((((((((....(((...)))..)))))))))........)). ( -20.30) >DroAna_CAF1 100387 99 + 1 UCCGUGGAUUGCGUGGCCGCUCCGCCCACGCA-UAA--------CGAGA------AAGUCCAAUAAAAUAUGUACAAAUAAUUUGCUAUGCGUAAUUUGUGCAAAUAAUACGCG (((((....(((((((.((...)).)))))))-..)--------)).))------...............(((((((((....(((...)))..)))))))))........... ( -24.80) >consensus UUC_UGCAUUGCGUG_________CCUACGCA_CAA________CGACA______AAAUCAAAUAAAAUAUGUACAAAUAAUUUGCCAUGCGUAAUUUGUGCAAAUAAUAUGCG .........((((((...........))))))......................................(((((((((....(((...)))..)))))))))........... (-13.39 = -12.92 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:16 2006