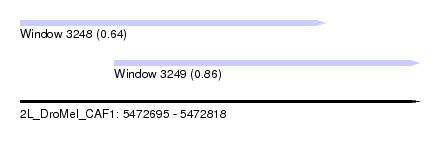
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,472,695 – 5,472,818 |
| Length | 123 |
| Max. P | 0.857410 |

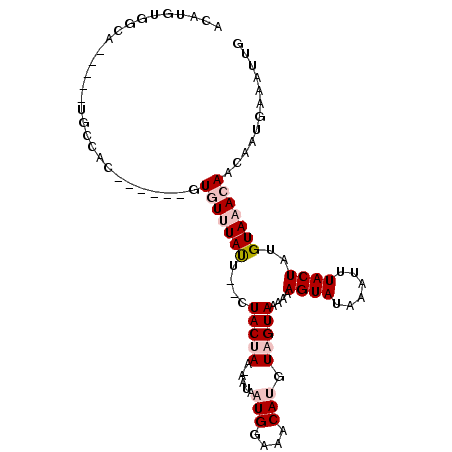
| Location | 5,472,695 – 5,472,789 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -13.83 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
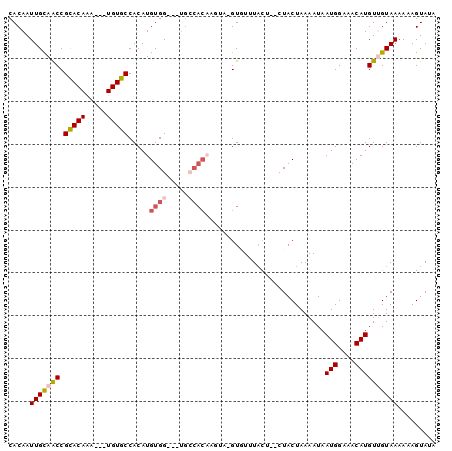
>2L_DroMel_CAF1 5472695 94 + 22407834 CACAAUUGCAACCGCACAAAAUUUGUGCCACAUGUGG---UGCCACAAGUA-GUGUUUACU--CUACUAAAAUAAUGGAAACAUGUUGUAAAAAAGUAUA .....(((((((.(((((.....)))))....((((.---...))))((((-(.(....).--)))))......(((....))))))))))......... ( -23.80) >DroPse_CAF1 109298 95 + 1 CACAAUUGUAACCGCACAAA---UGUGCCACAUGUGGCAGUGCCACAUCCCUGUUUAUAUA--CUACUAAAAUAAUGGAAACAUGUAGUAAAAAAGUAUA ......(((((.(((((...---.))))...(((((((...)))))))....).)))))((--((((.......(((....))))))))).......... ( -21.91) >DroSec_CAF1 91948 94 + 1 CACAAUUGCAACCGCACAAAAUUUGUGCCACAUGUGG---UGCCACAAGUA-GUAUUUCCU--CUACUAAAAUAAUGGAAACAUGUUGUAAAAAAGUAUA .....(((((((.(((((.....)))))....((((.---...))))((((-(........--)))))......(((....))))))))))......... ( -23.10) >DroEre_CAF1 93055 94 + 1 CACAAUUGCAGCCGUACAGAAUUUGUGCCACAUGUGG---UGGCACUAGUA-GUGUUUACU--CUACUAAAAUAAUGGAAACAUGUUGUAAAAAAGUAUA .....(((((((............(((((((.....)---))))))(((((-(.(....).--)))))).....(((....))))))))))......... ( -27.80) >DroAna_CAF1 99827 84 + 1 CACAAUUGCAACCGCACAAA---UGUGCCACAUGUGG---C---------U-GUGUUUACUCACUACUAAAAUAAUGGAAACAUGUUGUAAAAAAGUAUA .....(((((((.(((((..---...((((....)))---)---------)-))))..................(((....))))))))))......... ( -18.90) >DroPer_CAF1 110028 95 + 1 CACAAUUGUAACCGCACAAA---UGUGCCACAUGUGGCAGUGCCACAUCCAUGUUUAUAUA--CUACUAAAAUAAUGGAAACAUGUAGUAAAAAAGUAUA ......(((((.(((((...---.))))...(((((((...)))))))....).)))))((--((((.......(((....))))))))).......... ( -21.91) >consensus CACAAUUGCAACCGCACAAA___UGUGCCACAUGUGG___UGCCACAAGUA_GUGUUUACU__CUACUAAAAUAAUGGAAACAUGUUGUAAAAAAGUAUA .....(((((((.(((((.....))))).....((((.....))))............................(((....))))))))))......... (-13.83 = -14.50 + 0.67)

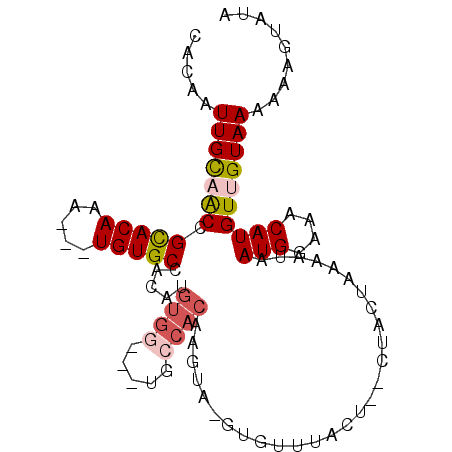
| Location | 5,472,724 – 5,472,818 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -8.85 |
| Energy contribution | -9.97 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5472724 94 + 22407834 ACAUGUGG-------UGCCACAAGUA-GUGUUUACU--CUACUAA-AAUAAUGGAAACAUGUUGUAAAAAAGUAUAAAUUUACUAUGUAAACAACAAUGAAAUUG ...((((.-------...))))((((-(.(....).--)))))..-....(((....)))(((((.....((((......))))......))))).......... ( -18.50) >DroVir_CAF1 135219 93 + 1 ACAUGUGGAAUGUUUUGCGAC-------UGUUUAUUAACUACUAAAAAUAAUGGAAACAAGUAGUAAA-AAGUAUAAAUUUACUAUGUAAACAACAAUGAG---- .((((..((....))..)...-------(((((((..((((((........((....))))))))...-.((((......))))..)))))))...)))..---- ( -15.10) >DroPse_CAF1 109324 98 + 1 ACAUGUGGCA----GUGCCACAUCCCUGUUUAUAUA--CUACUAA-AAUAAUGGAAACAUGUAGUAAAAAAGUAUAAAUUUACUAUGUAAACAACAAUGAAAUUG ..(((((((.----..)))))))...(((((((((.--.(((((.-....(((....))).)))))....((((......)))))))))))))............ ( -24.40) >DroMoj_CAF1 149735 93 + 1 ACAUGUGGCGUGUAUUGCGAC-------UGUUUAUUAACUACUAAAAAUAAUGGAAACAAGUAGUAAA-AAGUAUAAAUUUACUAUGUAAACAACAAUGAG---- .(((((.(((.....))).))-------(((((((..((((((........((....))))))))...-.((((......))))..)))))))...)))..---- ( -18.20) >DroAna_CAF1 99853 87 + 1 ACAUGUGG-------C---------U-GUGUUUACUCACUACUAA-AAUAAUGGAAACAUGUUGUAAAAAAGUAUAAAUUUACUAUGUAAACAACAACGAAAUUG ...(((..-------.---------.-.(((((((....(((.((-....(((....))).)))))....((((......))))..)))))))...)))...... ( -13.30) >DroPer_CAF1 110054 98 + 1 ACAUGUGGCA----GUGCCACAUCCAUGUUUAUAUA--CUACUAA-AAUAAUGGAAACAUGUAGUAAAAAAGUAUAAAUUUACUAUGUAAACAACAAUGAAAUUG ..(((((((.----..)))))))...(((((((((.--.(((((.-....(((....))).)))))....((((......)))))))))))))............ ( -24.60) >consensus ACAUGUGGCA_____UGCCAC______GUGUUUAUU__CUACUAA_AAUAAUGGAAACAUGUAGUAAAAAAGUAUAAAUUUACUAUGUAAACAACAAUGAAAUUG ............................(((((((....(((((......(((....))).)))))....((((......))))..)))))))............ ( -8.85 = -9.97 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:15 2006