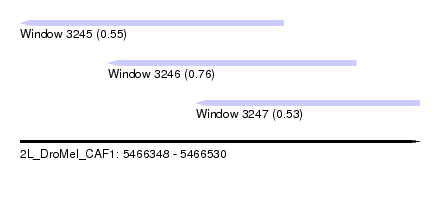
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,466,348 – 5,466,530 |
| Length | 182 |
| Max. P | 0.756653 |

| Location | 5,466,348 – 5,466,468 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -56.00 |
| Consensus MFE | -38.74 |
| Energy contribution | -37.88 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
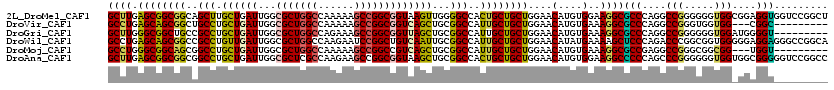
>2L_DroMel_CAF1 5466348 120 - 22407834 GCUUGAGCGGCGGCAGCUUGCUGAUUGGCGCUGGCCAAAAAGCCGGCGGUAAGUUGGGGCCACUGCUGCUGGAACAUGUGGAAGGCGCCCAGGCCGGGGGGUGGCGGAGGUGGUCCGGCU .....(((((.(((..((.(((...((.(((((((......))))))).))))).)).))).)))))((((((.(((.(....(.(((((........))))).)..).))).)))))). ( -52.70) >DroVir_CAF1 125454 108 - 1 GCCUGAGCAGCGGCUGCCUGCUGAUUGGCGCUGGCCAAAAAGCCGGCGGUCAGCUGCGGCCAUUGCUGCUGGAACAUGUGAAAGGCGCCCAGGCCGGGUGGUGG---CGGC--------- (((...((((((((((((.(((..(((((....)))))..))).))))))).))))).(((((..((((........))....(((......))).))..))))---))))--------- ( -56.40) >DroGri_CAF1 133292 111 - 1 GCUUGGGCGGCUGCCGCCUGCUGAUUGGCGCUGGCCAGAAAGCCGGCGGUUAGCUGCGGCCAUUGCUGCUGGAACAUGUGAAAGGCGCCCAGGCCGGGGGGUGGAUGGGGU--------- ((((((((((((((((((.(((..(((((....)))))..))).))))).)))).(((((....)))))................))))))))).................--------- ( -52.60) >DroWil_CAF1 127166 120 - 1 GCCUGAGCAGCGGCCGCCUGUUGAUUGGCGCUGGCCAAGAAUCCGGCUGUCAAUUGCGGCCAUUGCUGCUGGAACAUAUGAAAAGCUCCCAGACCCGGCGGUGGGGGAGGAGGGCCGGCA (((..((((((((((((.....((((((((((((........))))).)))))))))))))...))))))((..(...(....).(((((..(((....)))..)))))..)..))))). ( -55.30) >DroMoj_CAF1 139573 108 - 1 GCCUGGGCGGCAGCGGCCUGCUGAUUGGCGCUGGCCAAAAAGCCGGCCGUCAGCUGCGGCCAUUGCUGCUGGAACAUGUGAAAGGCGCCGAGGCCGGGCGGCGG---UGGU--------- ......(((((.((((((.(((..(((((....)))))..))).))))))..))))).((((((((((((((.....(((.....))).....)).))))))))---))))--------- ( -55.70) >DroAna_CAF1 91048 120 - 1 GCUUGAGCGGCGGCGGCCUGCUGAUUGGCGCUCGCCAAGAAGCCGGCGGUAAGCUGCGGCCACUGCUGCUGGAACAUGUGGAAGGCCCCCAGCCCGGGGGGUGGUGGCGGGGGUCCGGCC (.((.((((((((.((((.(((..(((((....)))))..)))..((((....)))))))).)))))))).)).)....((..(((((((.((((........).)))))))).))..)) ( -63.30) >consensus GCCUGAGCGGCGGCCGCCUGCUGAUUGGCGCUGGCCAAAAAGCCGGCGGUCAGCUGCGGCCAUUGCUGCUGGAACAUGUGAAAGGCGCCCAGGCCGGGGGGUGG_GGAGGU_________ ((((.((((((((..(((.((((((...(((((((......)))))))))))))...)))..))))))))....(....)..))))(((....(((.....)))....)))......... (-38.74 = -37.88 + -0.86)
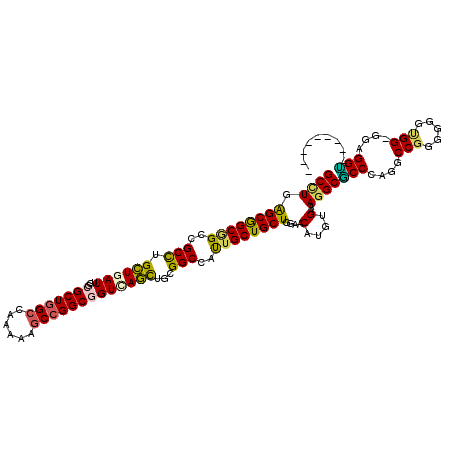
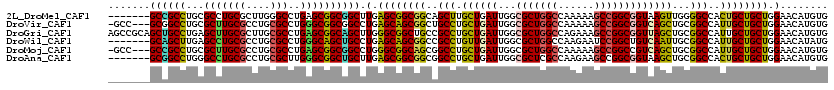
| Location | 5,466,388 – 5,466,501 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.21 |
| Mean single sequence MFE | -61.33 |
| Consensus MFE | -50.06 |
| Energy contribution | -49.15 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5466388 113 - 22407834 -------GCCGCCUGCGCCUGCGCUUGGGCCUGAGCGGCGGCUUGAGCGGCGGCAGCUUGCUGAUUGGCGCUGGCCAAAAAGCCGGCGGUAAGUUGGGGCCACUGCUGCUGGAACAUGUG -------((((((.((((((......))))....))))))))((.((((((((..(((..(..((((.(((((((......))))))).).)))..))))..)))))))).))....... ( -54.00) >DroVir_CAF1 125482 116 - 1 -GCC---GCGGCCUGCGCUUGCGCCUGCGCCUGGGCGGCGGCCUGAGCAGCGGCUGCCUGCUGAUUGGCGCUGGCCAAAAAGCCGGCGGUCAGCUGCGGCCAUUGCUGCUGGAACAUGUG -(((---((.((((((((........))))..)))).))))).(.((((((((((((..((((((...(((((((......))))))))))))).))))))...)))))).)........ ( -68.10) >DroGri_CAF1 133323 120 - 1 AGCCGCAGCUGCCUGAGCUUGCGCUUGCGCCUGAGCGGCAGCUUGGGCGGCUGCCGCCUGCUGAUUGGCGCUGGCCAGAAAGCCGGCGGUUAGCUGCGGCCAUUGCUGCUGGAACAUGUG .((((((((((((.((((....))))..(((..(((....)))..)))))).((((((.(((..(((((....)))))..))).)))))).))))))))).................... ( -64.60) >DroWil_CAF1 127206 113 - 1 -------GCAGCUUGAGCCUGCGCCUGCGCCUGGGCAGCUGCCUGAGCAGCGGCCGCCUGUUGAUUGGCGCUGGCCAAGAAUCCGGCUGUCAAUUGCGGCCAUUGCUGCUGGAACAUAUG -------((((((...(((((((....)))..)))))))))).(.((((((((((((.....((((((((((((........))))).)))))))))))))...)))))).)........ ( -53.40) >DroMoj_CAF1 139601 116 - 1 -GCC---GCCGCCUGCGCUUGCGCCUGCGCCUGAGCGGCGGCCUGGGCGGCAGCGGCCUGCUGAUUGGCGCUGGCCAAAAAGCCGGCCGUCAGCUGCGGCCAUUGCUGCUGGAACAUGUG -(((---((((((.((((........))))..).)))))))).(.((((((((.((((.((.(.(((((((((((......))))).))))))).)))))).)))))))).)........ ( -65.80) >DroAna_CAF1 91088 113 - 1 -------GCGGCCUGGGCCUGCGCCUGCGCUUGGGCGGCUGCUUGAGCGGCGGCGGCCUGCUGAUUGGCGCUCGCCAAGAAGCCGGCGGUAAGCUGCGGCCACUGCUGCUGGAACAUGUG -------((((((...(((((((....)))..))))))))))((.((((((((.((((.(((..(((((....)))))..)))..((((....)))))))).)))))))).))....... ( -62.10) >consensus _______GCCGCCUGCGCCUGCGCCUGCGCCUGAGCGGCGGCCUGAGCGGCGGCCGCCUGCUGAUUGGCGCUGGCCAAAAAGCCGGCGGUCAGCUGCGGCCAUUGCUGCUGGAACAUGUG .......((((((...(((((((....)))..)))))))))).(.((((((((..(((.((((((...(((((((......)))))))))))))...)))..)))))))).)........ (-50.06 = -49.15 + -0.91)

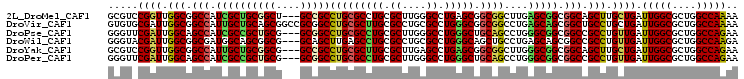
| Location | 5,466,428 – 5,466,530 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.27 |
| Mean single sequence MFE | -56.83 |
| Consensus MFE | -42.62 |
| Energy contribution | -41.90 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5466428 102 - 22407834 GCGUCCGGUUGGCGGCCAUCGCUGCGGCU---GCCGCCUGCGCCUGCGCUUGGGCCUGAGCGGCGGCUUGAGCGGCGGCAGCUUGCUGAUUGGCGCUGGCCAAAA ......(((..(((.(((...(.((((((---((((((.((....))(((..((((........))))..)))))))))))).))).)..))))))..))).... ( -59.50) >DroVir_CAF1 125522 105 - 1 GUGUGCGAUUGGCGGCCAUUGCUGCAGCGGCCGCGGCCUGCGCUUGCGCCUGCGCCUGGGCGGCGGCCUGAGCAGCGGCUGCCUGCUGAUUGGCGCUGGCCAAAA ....(((...(((((((...(((((...((((((.((((((((........))))..)))).))))))...)))))))))))))))...(((((....))))).. ( -59.60) >DroPse_CAF1 102358 102 - 1 GGGUUCGAUUGGCAGCCAUCGCCGCUGCG---GCGGCCUGCGCCUGCGCUUGGGCCUGGGCUGCAGCCUGGGCGGCGGCCGCCUGUUGAUUGGCGCUGGCCAGAA ....(((((.(((.(((..((((((((((---((((((((((....)))..)))))...))))))))...))))..))).))).)))))(((((....))))).. ( -57.30) >DroWil_CAF1 127246 102 - 1 GGGUACGAUUGGCGGCGAUGGCAGCGGCG---GCAGCUUGAGCCUGCGCCUGCGCCUGGGCAGCUGCCUGAGCAGCGGCCGCCUGUUGAUUGGCGCUGGCCAAGA ........(((((((((....((((((((---((.......(((((((....)))..)))).(((((....))))).)))))).)))).....)))).))))).. ( -50.10) >DroYak_CAF1 93762 102 - 1 GCGUCCGGUUGGCGGCCAUUGCUGCGGCG---GCCGCCUGCGCUUGCGCUUGAGCCUGAGCGGCGGCUUGGGCGGCGGCAGCUUGCUGAUUGGCGCUGGCCAGAA ...((.(((..(((.(((...(.(((((.---((((((.(((....)))....(((..(((....)))..))))))))).)).))).)..))))))..))).)). ( -57.20) >DroPer_CAF1 103075 102 - 1 GGGUUCGAUUGGCAGCCAUCGCCGCUGCG---GCGGCCUGCGCCUGCGCUUGGGCCUGGGCUGCAGCCUGGGCGGCGGCCGCCUGUUGAUUGGCGCUGGCCAGAA ....(((((.(((.(((..((((((((((---((((((((((....)))..)))))...))))))))...))))..))).))).)))))(((((....))))).. ( -57.30) >consensus GGGUCCGAUUGGCGGCCAUCGCUGCGGCG___GCGGCCUGCGCCUGCGCUUGGGCCUGGGCGGCGGCCUGAGCGGCGGCCGCCUGCUGAUUGGCGCUGGCCAAAA .....((((.(((.(((.((((((((((....)))))(((((((((.((....)).))))).))))....))))).))).))).)))).(((((....))))).. (-42.62 = -41.90 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:13 2006