| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,461,982 – 5,462,090 |
| Length | 108 |
| Max. P | 0.655601 |

| Location | 5,461,982 – 5,462,090 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -46.67 |
| Consensus MFE | -28.21 |
| Energy contribution | -28.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
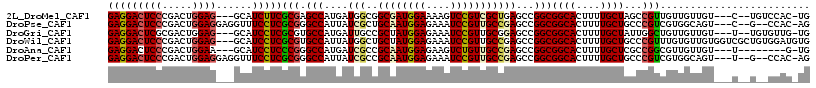
>2L_DroMel_CAF1 5461982 108 - 22407834 GAGGACUCCCGACUGGAG---GCAUCUUCGCGAGCCAUGAUGGCGGCGAUGGAAAAGUCCGUCGCUGAGCCGGCGGCACUUUUGCUAGCCGUUGUUGUUGU---C--UGUCCAC-UG ..((((...((((....(---((.((.....)))))..((..(((((...(.((((((((((((......)))))).)))))).)..)))))..)))))).---.--.))))..-.. ( -44.40) >DroPse_CAF1 97457 109 - 1 GAGGACUCCCGACUGGAGGAGGUUUCCUCGCGGGCCAUUAUCGCUGCAAUGGAGAAAUCCGUUGCCGAGCCGGCGGCACUUUUGCUGCCCGUCGUGGCAGU---C--G--CCAC-AG ((((((((((.....).))))...)))))((((((.....(((..((((((((....))))))))))))))(((((((....)))))))))).(((((...---.--)--))))-.. ( -51.90) >DroGri_CAF1 127443 108 - 1 GAGGACUCGCGACUGGAG---GCAUCCUCGCGUGCCAUGAUUGCCGCUAUGGAGAAAUCCGUUGCGGAGCCGGCGGCACUUUUGCUAUUGGCUGUUGUUGU---U--UGUGUUG-UG ..((.(.(((((..(((.---...)))))))).))).......((((.(((((....))))).))))((((((..(((....)))..))))))........---.--.......-.. ( -41.20) >DroWil_CAF1 123450 114 - 1 GAGGACUCCCGACUGGAG---GCAUCCUCGCGUGCCAUUAUGGCUGCUAUGGAGAAAUCCGUUGCCGAGCCGGCGGCACUUUUGCUGCCCGUUUGUGUUGUGGUCGCUGUGGAUGUG .....((((.....))))---((((((.((((.(((((.((((((((.(((((....))))).))..))))(((((((....))))))).......)).)))))))).).)))))). ( -50.90) >DroAna_CAF1 87241 102 - 1 GAGGACUCCCGACUGGAA---GCAUCCUCCCGGGCCAUGAUCGCCGCAAUGGAGAAGUCUGUUGCCGAGCCGGCGGCACUUUUGCUCGCCGGCGUUGUUGU---U--------G-UG ((.(((.((((...(((.---...)))...))))......(((..((((((((....)))))))))))((((((((((....))).)))))))))).))..---.--------.-.. ( -39.40) >DroPer_CAF1 98148 109 - 1 GAGGACUCCCGACUGGAGGAGGUUUCCUCGCGGGCCAUUAUCGCCGCAAUGGAGAAAUCCGUUGCCGAGCCGGCGGCACUUUUGCUGCCCGUCGUGGCAGU---U--G--CCAC-AG ((((((((((.....).))))...)))))((((((.....(((..((((((((....))))))))))))))(((((((....)))))))))).(((((...---.--)--))))-.. ( -52.20) >consensus GAGGACUCCCGACUGGAG___GCAUCCUCGCGGGCCAUGAUCGCCGCAAUGGAGAAAUCCGUUGCCGAGCCGGCGGCACUUUUGCUGCCCGUCGUUGUUGU___C__U__CCAC_UG (((((((((.....))))......)))))(((.(((....(((..((((((((....)))))))))))...)))((((....))))...)))......................... (-28.21 = -28.52 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:10 2006