
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,453,966 – 5,454,072 |
| Length | 106 |
| Max. P | 0.931746 |

| Location | 5,453,966 – 5,454,072 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -16.18 |
| Energy contribution | -17.26 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5453966 106 + 22407834 CAAAAA---GAAACGA------GAAAAGGGGGAAAAUUCUUGAGGGGAGUGCCGCCAAGAAAGGCGGGAAAGGAGAAAGAGCUAAGUUAUUACAUUUAGCGGGCACUAAAAUGUC ......---....(((------(((...........)))))).....(((((((((......)))...............(((((((......))))))).))))))........ ( -25.30) >DroSec_CAF1 73375 99 + 1 ---AAA---GAAACGU------GAAAAGGGGGAAAAUUCUUGAGGGGAGUGCCGCCAAGAAAGGCGGGAAAGGA----GAGCUAAGUUAUUAGAUUUAGCGGGCAAUAAAAUGUC ---...---...((((------.............(((((.....)))))((((((......))).........----..((((((((....)))))))).)))......)))). ( -20.90) >DroSim_CAF1 84497 105 + 1 G-AAAA---GAAACGU------GAAAAGGGGGAAAAUUCUUGAGGGGAGUGCCGCCAAGAAAGGCGGGAAAGGAGAAAGAGCUAAGUUAUUACAUUUAGCGGGCACUAAAAUGUC .-....---....(.(------...(((((......)))))..).).(((((((((......)))...............(((((((......))))))).))))))........ ( -23.70) >DroEre_CAF1 74439 111 + 1 G-AAAAUAAACGAGGCGCAGGGGGAAAAGGGGAAAAUUCUUGAAGGGAGUGCC---ACGAAAGGCGGGAAAGGAAAAAGAGCUAAGUUAUUACAUUUAGCGGGCACUAAAAUGUC .-...........((((((((((.............)))))).....((((((---.(....).................(((((((......))))))).))))))....)))) ( -22.02) >DroYak_CAF1 81000 108 + 1 G-AAAAUAAGAAACGU---GAGGAAAAGGGGGAAAAUUCCUGAAGGGAGUGCC---ACGAAAGGCGGGAAAGGAAAAAGAGCUAAGUUAUUACAUUUAGCGGGCACUAAAAUGUC .-...........(..---..)....(((((.....)))))......((((((---.(....).................(((((((......))))))).))))))........ ( -23.10) >consensus G_AAAA___GAAACGU______GAAAAGGGGGAAAAUUCUUGAGGGGAGUGCCGCCAAGAAAGGCGGGAAAGGAAAAAGAGCUAAGUUAUUACAUUUAGCGGGCACUAAAAUGUC ..........................(((((.....)))))......(((((((((......)))...............(((((((......))))))).))))))........ (-16.18 = -17.26 + 1.08)



| Location | 5,453,966 – 5,454,072 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -15.86 |
| Consensus MFE | -11.10 |
| Energy contribution | -12.50 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5453966 106 - 22407834 GACAUUUUAGUGCCCGCUAAAUGUAAUAACUUAGCUCUUUCUCCUUUCCCGCCUUUCUUGGCGGCACUCCCCUCAAGAAUUUUCCCCCUUUUC------UCGUUUC---UUUUUG (((.....((((((.(((((.(......).)))))...............(((......))))))))).......((((...........)))------).)))..---...... ( -18.50) >DroSec_CAF1 73375 99 - 1 GACAUUUUAUUGCCCGCUAAAUCUAAUAACUUAGCUC----UCCUUUCCCGCCUUUCUUGGCGGCACUCCCCUCAAGAAUUUUCCCCCUUUUC------ACGUUUC---UUU--- (((.......((((.(((((..........)))))..----.........(((......)))))))..........(((...........)))------..)))..---...--- ( -14.20) >DroSim_CAF1 84497 105 - 1 GACAUUUUAGUGCCCGCUAAAUGUAAUAACUUAGCUCUUUCUCCUUUCCCGCCUUUCUUGGCGGCACUCCCCUCAAGAAUUUUCCCCCUUUUC------ACGUUUC---UUUU-C (((.....((((((.(((((.(......).)))))...............(((......)))))))))........(((...........)))------..)))..---....-. ( -18.40) >DroEre_CAF1 74439 111 - 1 GACAUUUUAGUGCCCGCUAAAUGUAAUAACUUAGCUCUUUUUCCUUUCCCGCCUUUCGU---GGCACUCCCUUCAAGAAUUUUCCCCUUUUCCCCCUGCGCCUCGUUUAUUUU-C .........((((..(((((.(......).)))))...............(((......---)))................................))))............-. ( -13.30) >DroYak_CAF1 81000 108 - 1 GACAUUUUAGUGCCCGCUAAAUGUAAUAACUUAGCUCUUUUUCCUUUCCCGCCUUUCGU---GGCACUCCCUUCAGGAAUUUUCCCCCUUUUCCUC---ACGUUUCUUAUUUU-C .((((((.(((....)))))))))(((((...(((...............(((......---))).........(((((...........))))).---..)))..)))))..-. ( -14.90) >consensus GACAUUUUAGUGCCCGCUAAAUGUAAUAACUUAGCUCUUUCUCCUUUCCCGCCUUUCUUGGCGGCACUCCCCUCAAGAAUUUUCCCCCUUUUC______ACGUUUC___UUUU_C ........((((((.(((((.(......).)))))...............(((......)))))))))............................................... (-11.10 = -12.50 + 1.40)

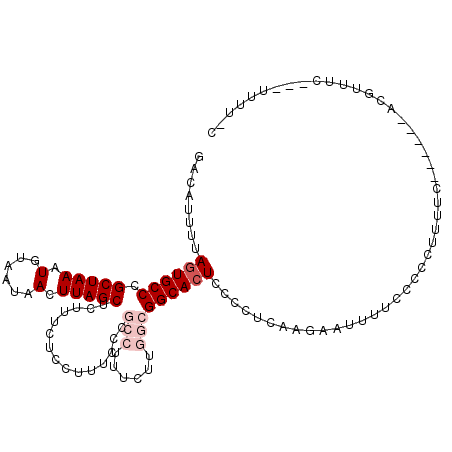
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:08 2006