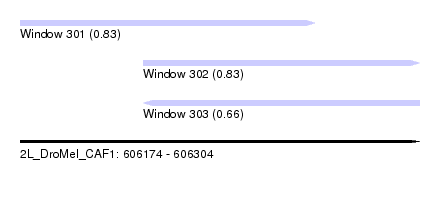
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 606,174 – 606,304 |
| Length | 130 |
| Max. P | 0.831661 |

| Location | 606,174 – 606,270 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -21.39 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 606174 96 + 22407834 CCGACAACAAACUGCGCCUGCGCACGCGUUCAAUUAGCUGCAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGC------------------------GAUGUCGCCACAAAC .(((((......(((((.(((((.((((((.....)))(((.....)))......)))))))).))))).(((......))------------------------).)))))........ ( -32.30) >DroVir_CAF1 47824 85 + 1 UCAGGUACAAACUGCGGCAGUGCACGCGUUCAAUUAGCUGCAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCC------------------------------GCUGUC-----CAGU ..........((((.(((((((.((((((.......((((((...(....)...))))))....))))...)).)------------------------------))))))-----)))) ( -27.60) >DroEre_CAF1 38051 96 + 1 CCGACAACAAACUGCGCCUGCGCACGCGUUCAAUUAGCUGCAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGC------------------------GAUGUCGCCACAAAC .(((((......(((((.(((((.((((((.....)))(((.....)))......)))))))).))))).(((......))------------------------).)))))........ ( -32.30) >DroYak_CAF1 38068 96 + 1 CCGACAACAAACUGCGCCUGCGCACGCGUUCAAUUAGCUGCAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGC------------------------GAUGUCGCCACAAAC .(((((......(((((.(((((.((((((.....)))(((.....)))......)))))))).))))).(((......))------------------------).)))))........ ( -32.30) >DroMoj_CAF1 46860 85 + 1 UCAGCUACAAACUGCGAAAGCACACGCGUUCAAUUAGCUGCAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCC------------------------------GCUGUC-----CAGU .((((.......(((....)))...((((.......((((((...(....)...))))))....)))).......------------------------------))))..-----.... ( -24.30) >DroAna_CAF1 37053 120 + 1 UCGAUAACUAACUGCGCCUGCGCACGCGUUCAAUUAGCUGCAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGGUGCCCGGUUCGGUGCUGGUGCCGGGCUGAUGUCGCCACAAAC .(((((......(((((.(((((.((((((.....)))(((.....)))......)))))))).))))).((.(((((..((.(((.....)))))..))))).)).)))))........ ( -48.90) >consensus CCGACAACAAACUGCGCCUGCGCACGCGUUCAAUUAGCUGCAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGC________________________GAUGUCGCCACAAAC ............(((((.(((((.((((((.....)))(((.....)))......)))))))).)))))................................................... (-21.39 = -22.08 + 0.69)

| Location | 606,214 – 606,304 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -16.39 |
| Energy contribution | -17.95 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
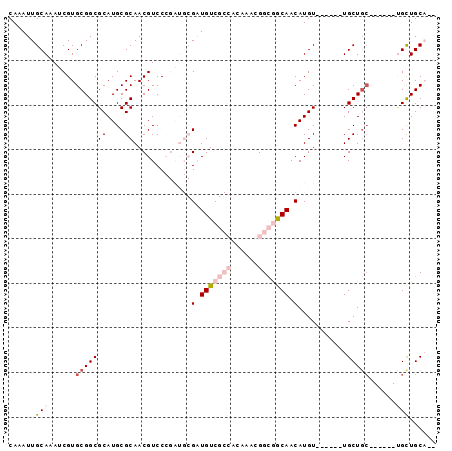
>2L_DroMel_CAF1 606214 90 + 22407834 CAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGCGAUGUCGCCACAAACGGCGGCAACAUGU------UGCUGCUGCUGCUGCUGCAAC ....(((((......((((((((.(((((((((......))(.(((((((......))))))).)..))------))).))))).)))))))))). ( -35.60) >DroVir_CAF1 47864 82 + 1 CAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCC------GCUGUC-----CAGUGGCAACAACAUGUAGCUUUUGCUGCUGCUGCCGUUGC--- ......((((..((.(((((((....))...(.((------(((...-----.))))))........(((((....))))).)))))))))))--- ( -26.40) >DroSec_CAF1 37651 81 + 1 CAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGCGAUGUCGCCACAAACGGCGGCAACAUGU------UGCU---------GCUGCAAC ....(((((......((((((((((((((.((...)).)))).(((((((......))))))).)))).------))))---------))))))). ( -32.60) >DroEre_CAF1 38091 82 + 1 CAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGCGAUGUCGCCACAAACGGCGGCAACAUGU------UGCUGC------UGCUGCA-- .....((((.....((.(((((((.((((.((...)).))))))).)))).))..(((((((((....)------))))))------)).))))-- ( -33.70) >DroYak_CAF1 38108 84 + 1 CAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGCGAUGUCGCCACAAACGGCGGCAACAUGU------UGCUGC------UGCUGCAAC ....(((((.....((.(((((((.((((.((...)).))))))).)))).))..(((((((((....)------))))))------)).))))). ( -35.20) >DroMoj_CAF1 46900 76 + 1 CAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCC------GCUGUC-----CAGUGGUAACAACAUGU------UGCUGCCGCUGCUGUUGC--- ......((((.....((((((...(((((((((((------(((...-----.))))).......))))------))).))))))))..))))--- ( -26.51) >consensus CAAAUUGCAAAUCGUGCGGCGCAUGCGCAACGUCCCGAUGCGAUGUCGCCACAAACGGCGGCAACAUGU______UGCUGC______UGCUGCA__ ......(((......(((((((....)).............(.(((((((......))))))).)...........)))))......)))...... (-16.39 = -17.95 + 1.56)

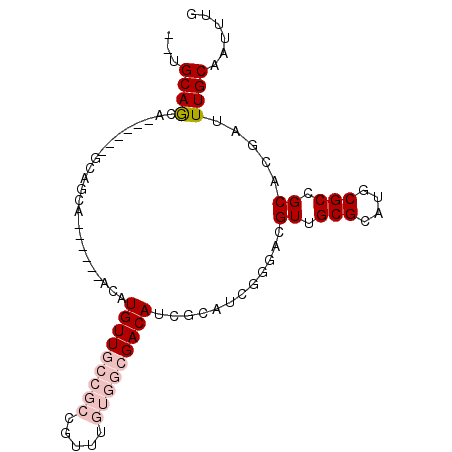
| Location | 606,214 – 606,304 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -17.06 |
| Energy contribution | -18.50 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
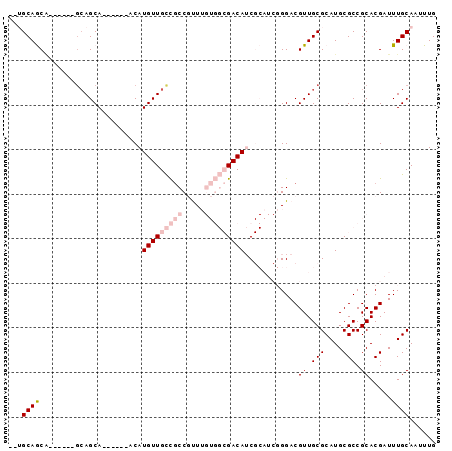
>2L_DroMel_CAF1 606214 90 - 22407834 GUUGCAGCAGCAGCAGCAGCA------ACAUGUUGCCGCCGUUUGUGGCGACAUCGCAUCGGGACGUUGCGCAUGCGCCGCACGAUUUGCAAUUUG ((((((((.((.((((((...------...)))))).)).)).(((((((.(((((((.((...)).)))).)))))))))).....))))))... ( -37.10) >DroVir_CAF1 47864 82 - 1 ---GCAACGGCAGCAGCAGCAAAAGCUACAUGUUGUUGCCACUG-----GACAGC------GGACGUUGCGCAUGCGCCGCACGAUUUGCAAUUUG ---((((.((((((((((((....))....))))))))))....-----....((------((.(((.......))))))).....))))...... ( -30.50) >DroSec_CAF1 37651 81 - 1 GUUGCAGC---------AGCA------ACAUGUUGCCGCCGUUUGUGGCGACAUCGCAUCGGGACGUUGCGCAUGCGCCGCACGAUUUGCAAUUUG ((((((((---------.(((------(....)))).))(((..((((((.(((((((.((...)).)))).))))))))))))...))))))... ( -32.40) >DroEre_CAF1 38091 82 - 1 --UGCAGCA------GCAGCA------ACAUGUUGCCGCCGUUUGUGGCGACAUCGCAUCGGGACGUUGCGCAUGCGCCGCACGAUUUGCAAUUUG --((((((.------(((((.------....))))).))(((..((((((.(((((((.((...)).)))).))))))))))))...))))..... ( -33.30) >DroYak_CAF1 38108 84 - 1 GUUGCAGCA------GCAGCA------ACAUGUUGCCGCCGUUUGUGGCGACAUCGCAUCGGGACGUUGCGCAUGCGCCGCACGAUUUGCAAUUUG ((((((((.------(((((.------....))))).))(((..((((((.(((((((.((...)).)))).))))))))))))...))))))... ( -34.90) >DroMoj_CAF1 46900 76 - 1 ---GCAACAGCAGCGGCAGCA------ACAUGUUGUUACCACUG-----GACAGC------GGACGUUGCGCAUGCGCCGCACGAUUUGCAAUUUG ---((((.....(((((.(((------((.(((((((.......-----))))))------)...)))))(....)))))).....))))...... ( -26.60) >consensus __UGCAGCA______GCAGCA______ACAUGUUGCCGCCGUUUGUGGCGACAUCGCAUCGGGACGUUGCGCAUGCGCCGCACGAUUUGCAAUUUG ...((((.......................(((((((((.....)))))))))............((.(((....))).)).....))))...... (-17.06 = -18.50 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:09 2006