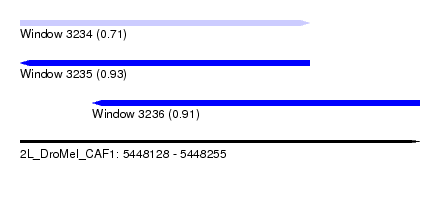
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,448,128 – 5,448,255 |
| Length | 127 |
| Max. P | 0.934480 |

| Location | 5,448,128 – 5,448,220 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -11.36 |
| Energy contribution | -11.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5448128 92 + 22407834 UAAAAAAAGAC-CGCGUCCUUCUUGGGUUUAUUUUAUU-----UUUUUUUUGUGUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACU .....(((((.-.((((((.....)))...........-----........((....))...)))...)))))...(((((((.....)))))))... ( -17.20) >DroPse_CAF1 79902 87 + 1 UAAAAAAAAACUCGCGUCCUGCUUGGGUUUUU---GUU-----UUGUUUU---UUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACU ((((((((((((((.(.....).)))))))))---.))-----)))....---....((....))...........(((((((.....)))))))... ( -18.60) >DroSec_CAF1 67686 93 + 1 UAAAAAAAGAC-CGCGUCCUUCUUGGGUUUAUUU-A---UUUUUUUUUUUCGUGUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACU .....(((((.-.((((((.....))).......-.---............((....))...)))...)))))...(((((((.....)))))))... ( -17.50) >DroEre_CAF1 68597 89 + 1 UAAA-AAAGUC-CGCGUCCUUCUUGGGUUUAUUUUAUUUU-------UUUUGUGUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACU ....-((((..-.((((((.....))).............-------....((....))...)))....))))...(((((((.....)))))))... ( -15.80) >DroWil_CAF1 98883 88 + 1 UACG-AAAACU-CGGAUACUGCUUACAUUUUUUUCC-------CCGUUUUCGU-UUCGUUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACU .(((-(((((.-.(((.................)))-------..))))))))-......................(((((((.....)))))))... ( -20.63) >DroPer_CAF1 79484 87 + 1 UAAAAAAAAACUCGCGUCCUGCUUGGGUUUUU---GUU-----UUGUUUU---UUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACU ((((((((((((((.(.....).)))))))))---.))-----)))....---....((....))...........(((((((.....)))))))... ( -18.60) >consensus UAAAAAAAAAC_CGCGUCCUGCUUGGGUUUAUUU_AUU_____UUGUUUU_GUGUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACU ................(((.....)))..............................((....))...........(((((((.....)))))))... (-11.36 = -11.42 + 0.06)

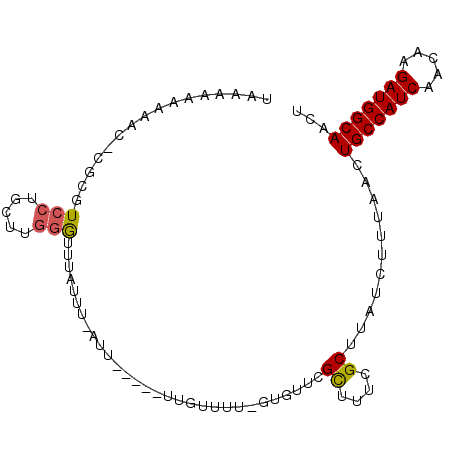
| Location | 5,448,128 – 5,448,220 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5448128 92 - 22407834 AGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAAAAAA-----AAUAAAAUAAACCCAAGAAGGACGCG-GUCUUUUUUUA ..((((((((.....))))))))..........((....))..............-----..............(((((((((...-))))))))).. ( -21.80) >DroPse_CAF1 79902 87 - 1 AGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAAA---AAAACAA-----AAC---AAAAACCCAAGCAGGACGCGAGUUUUUUUUUA ..((((((((.....))))))))..........((....))((((---((((...-----...---......((.....))((....)))))))))). ( -21.30) >DroSec_CAF1 67686 93 - 1 AGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACGAAAAAAAAAAA---U-AAAUAAACCCAAGAAGGACGCG-GUCUUUUUUUA ..((((((((.....))))))))..........((....))..................---.-..........(((((((((...-))))))))).. ( -21.80) >DroEre_CAF1 68597 89 - 1 AGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAA-------AAAAUAAAAUAAACCCAAGAAGGACGCG-GACUUU-UUUA ..((((((((.....))))))))..........((....))..........-------................((((((......-..))))-)).. ( -19.10) >DroWil_CAF1 98883 88 - 1 AGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAACGAA-ACGAAAACGG-------GGAAAAAAAUGUAAGCAGUAUCCG-AGUUUU-CGUA ..((((((((.....)))))))).....................-((((((((..-------(((.(....((....)).).))).-.)))))-))). ( -24.30) >DroPer_CAF1 79484 87 - 1 AGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAAA---AAAACAA-----AAC---AAAAACCCAAGCAGGACGCGAGUUUUUUUUUA ..((((((((.....))))))))..........((....))((((---((((...-----...---......((.....))((....)))))))))). ( -21.30) >consensus AGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACAC_AAAACAA_____AAU_AAAAAAACCCAAGAAGGACGCG_GUCUUUUUUUA ..((((((((.....))))))))..........((....)).....................................(((((....)))))...... (-15.78 = -16.00 + 0.22)

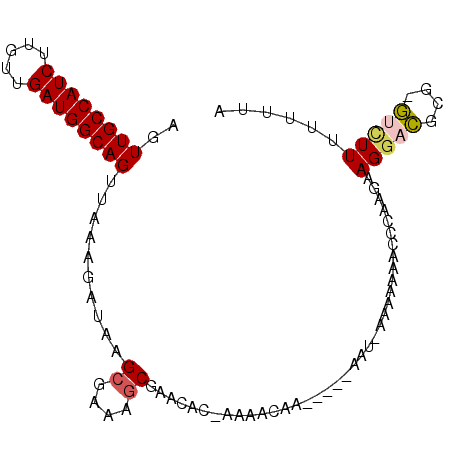
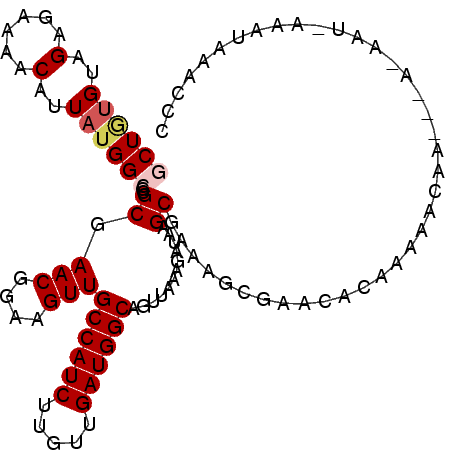
| Location | 5,448,151 – 5,448,255 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5448151 104 - 22407834 GCUGUGUAGGGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAAAAAA-----AAUAAAAUAAACCC ..(((((.............((((.....(((....)))((((((.....)))))).)))).......((....))..))))).......-----.............. ( -25.40) >DroGri_CAF1 105513 95 - 1 UCUAUGUAGAGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCACAAAAAAAGCUAAAAACAAA--AAAA------------ ....................((((.....(((....)))((((((.....)))))).......................))))........--....------------ ( -18.10) >DroSec_CAF1 67709 105 - 1 GCUGUGUAGGGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACGAAAAAAAAAAA---U-AAAUAAACCC ..(((((.............((((.....(((....)))((((((.....)))))).)))).......((....))..)))))...........---.-.......... ( -24.70) >DroEre_CAF1 68619 102 - 1 GCUGUGUAGGGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAA-------AAAAUAAAAUAAACCC ..(((((.............((((.....(((....)))((((((.....)))))).)))).......((....))..)))))...-------................ ( -25.40) >DroMoj_CAF1 110432 103 - 1 UCUAUGUAGAGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCACACAAAA---UAAAAACAAC--AAAA-AAAGUAAAUAC .................((((.(.(.((.(((....)))((((((.....))))))............)).).)...)---))).......--....-........... ( -17.90) >DroPer_CAF1 79508 96 - 1 GCU--GCAGAGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAAA---AAAACAA-----AAC---AAAAACCC (((--((......((((..((((((.....((....))))))))..))))....))))).........((....))....---.......-----...---........ ( -24.50) >consensus GCUGUGUAGAGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAAACAA___A_AAU_AAAUAAACCC ((((((..(......)..))))))..((.(((....)))((((((.....))))))............))....................................... (-17.70 = -18.20 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:03 2006