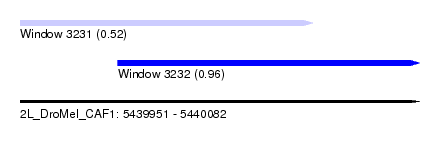
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,439,951 – 5,440,082 |
| Length | 131 |
| Max. P | 0.957978 |

| Location | 5,439,951 – 5,440,047 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -18.27 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
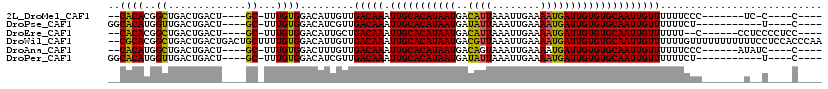
>2L_DroMel_CAF1 5439951 96 + 22407834 --CACACGGCUGACUGACU----GC-UUUGUGGACAUUGUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCC-------UC-C----C---- --.(((.(((.........----))-).)))(((.......(((((.(((((((((((..((((........)))))))))))))))))))).......-------))-)----.---- ( -23.84) >DroPse_CAF1 69713 95 + 1 GGCACAUGGUUGACUGACU----GC-UUUGUGGACAUCGUUGACAAAUUGCACAUAAUGAUAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCU-----------U----C---- ........((..((.((.(----((-(....)).))))))..)).(((((((((((((.((.((......)).)).))))))))))))).........-----------.----.---- ( -21.90) >DroEre_CAF1 60330 100 + 1 --CACACGGCUGACUGACU----GC-UUUGUGGACAUUGCUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUU--C------CCUCCCCUCC---- --.(((.(((.........----))-).)))(((.......(((((.(((((((((((..((((........))))))))))))))))))))....--.------..))).....---- ( -23.84) >DroWil_CAF1 84985 117 + 1 --CGCACGGCUGACUGACUGACUGCUUUUGUGGACAUUGUUGACAAAUUGCACAUAAUGACGUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUGUUUUUUUUUUUCCUCCACCCAA --.(((((((.....).)))..)))....(((((((....))...(((((((((((((..((((........))))))))))))))))).....................))))).... ( -23.70) >DroAna_CAF1 64072 98 + 1 --CACAUGGCUGACUGACU----GC-UUUGUGGACUUUGUUGACAAAUUGCACAUAAUGACAGUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCC------AUAUC----C---- --((((.(((.........----))-).)))).........(((((.(((((((((((.((((....)))....).)))))))))))))))).......------.....----.---- ( -21.60) >DroPer_CAF1 69351 95 + 1 GGCACAUGGUUGACUGACU----GC-UUUGUGGACAUCGUUGACAAAUUGCACAUAAUGAUAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCU-----------U----C---- ........((..((.((.(----((-(....)).))))))..)).(((((((((((((.((.((......)).)).))))))))))))).........-----------.----.---- ( -21.90) >consensus __CACACGGCUGACUGACU____GC_UUUGUGGACAUUGUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCUC_______U__C____C____ ..((((..((.............))...)))).........(((((.(((((((((((..((((........))))))))))))))))))))........................... (-18.27 = -17.72 + -0.55)

| Location | 5,439,983 – 5,440,082 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -17.08 |
| Consensus MFE | -15.87 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5439983 99 + 22407834 UUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCC-UC-C----CCCACCCCCAGUCAGUUUCUAUACCCAU-C--UCUUUU-G (((((.(((((((((((((..((((........)))))))))))))))))(......)..-..-.----..........))))).............-.--......-. ( -17.00) >DroSec_CAF1 59503 99 + 1 UUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUCC-CG-C----CCCUCCCCCAGUCAGCUUCUAUACCCAU-C--UCUUUU-G (((((.(((((((((((((..((((........)))))))))))))))))((........-.)-)----..........))))).............-.--......-. ( -17.00) >DroSim_CAF1 70535 101 + 1 UUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUUCAUGCC----CCCUCCCCCAGUCAGCUUCUAUACCCAU-C--UCUUUU-G (((((.(((((((((((((..((((........)))))))))))))))))(........).....----..........))))).............-.--......-. ( -17.60) >DroYak_CAF1 66631 106 + 1 UUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUCCCCUCCCCACCCCUCCUCCAGUCAGCUUCUAUACCCAU-C--UCAUACAG ..(((((.(((((((((((..((((........))))))))))))))))))))............................................-.--........ ( -16.50) >DroAna_CAF1 64104 98 + 1 UUGACAAAUUGCACAUAAUGACAGUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCCAUAUC----CCAUCC-CGCUUUAGC--CAAUACCUGU-C--UCUUUC-A ..(((((((((((((((((.((((....)))....).)))))))))))))...............----......-.((....))--.......)))-)--......-. ( -17.70) >DroPer_CAF1 69385 99 + 1 UUGACAAAUUGCACAUAAUGAUAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCU-----U----CCCUCCCUCACUCGCUGUCUCUAUCUAUUCUUUUUUUU-U ..(((((.(((((((((((.((.((......)).)).))))))))))))))))......-----.----......................................-. ( -16.70) >consensus UUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCC_UG_C____CCCUCCCCCAGUCAGCUUCUAUACCCAU_C__UCUUUU_G ..(((((.(((((((((((..((((........))))))))))))))))))))........................................................ (-15.87 = -15.90 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:59 2006