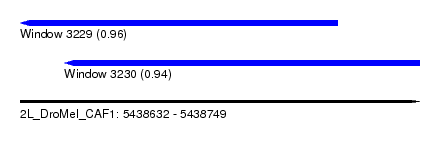
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,438,632 – 5,438,749 |
| Length | 117 |
| Max. P | 0.963961 |

| Location | 5,438,632 – 5,438,725 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
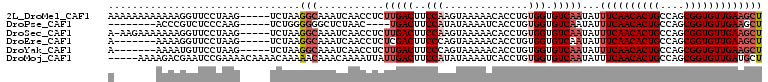
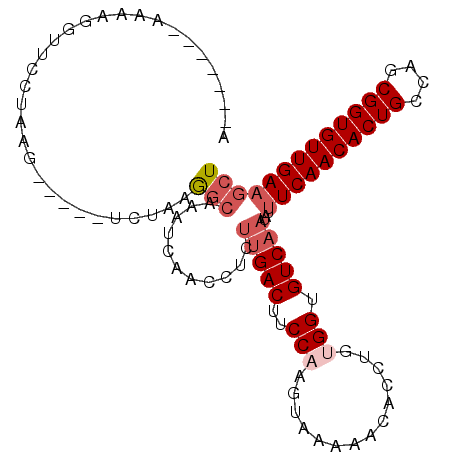
>2L_DroMel_CAF1 5438632 93 - 22407834 UAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC---------UCUGUCUCCGAU ..(((((..........(((((..(((.((......))...))).)))))..(((((((((((....)))))))))))...---------..)))))..... ( -24.70) >DroPse_CAF1 68188 96 - 1 UGGGGGGCUCUAAC----UGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAG--AAUCUGUCUCUGGA ....((((......----((((..(((((..........))))).))))....((((((((((....)))))))))))))).((((--(.......))))). ( -29.90) >DroGri_CAF1 92130 102 - 1 AAACGUAAAUAAAAUUAUUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAUGCUCAUCAGCUGAAGUGUCUCUGCU ....(((........(((((((..(((((..........))))).)))))))..(((((((((....)))))))))))).....(((.((......)).))) ( -26.60) >DroMoj_CAF1 94729 102 - 1 AAAAACAAACAAAAUUAUUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAUGCUCAUCAGCUGAAGUGUCUCUGCU ...............(((((((..(((((..........))))).)))))))..(((((((((....)))))))))........(((.((......)).))) ( -25.60) >DroAna_CAF1 62957 92 - 1 UAA-GCCAACUGACCUCUCGACUUUCCUCCGAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC---------UCUGUCUCCGGU ...-(((....(((.....(((...((.(.(.......).).)).)))....(((((((((((....)))))))))))...---------...)))...))) ( -22.50) >DroPer_CAF1 67851 96 - 1 UGGGGGGCUCUAAC----UGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAG--AAUCUGUCUCUGGA ....((((......----((((..(((((..........))))).))))....((((((((((....)))))))))))))).((((--(.......))))). ( -29.90) >consensus UAAGGCAAACUAAC_U_UUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAG___AUCUGUCUCUGGU ...................(((.............((((...))))......(((((((((((....)))))))))))...............)))...... (-18.28 = -18.62 + 0.33)

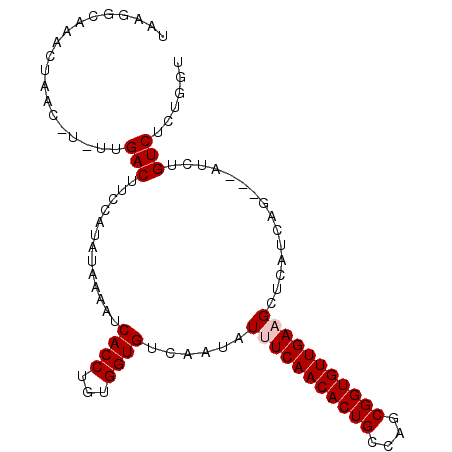
| Location | 5,438,645 – 5,438,749 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.99 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5438645 104 - 22407834 AAAAAAAAAAAAGGUUCCUAAG-----UCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCU ............((.....(((-----((..(((........)))...)))))))........(((((...))))).....(((((((((((....))))))))))).. ( -24.90) >DroPse_CAF1 68208 92 - 1 --------ACCCGUCUCCCAAG-----UCUGGGGGGCUCUAAC----UGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCU --------....((((((((..-----..))))))))......----((((..(((((..........))))).))))...(((((((((((....))))))))))).. ( -34.20) >DroSec_CAF1 58218 103 - 1 A-AAGAAAAAAAGGUUCCUAAG-----UCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCU .-..........((.....(((-----((..(((........)))...)))))))........(((((...))))).....(((((((((((....))))))))))).. ( -24.90) >DroEre_CAF1 58981 97 - 1 A-------AAAAGGUUCCUAAG-----UCUAAGGCAAAUCAACCUCUCGACUUCCCAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCU .-------...(((((.....(-----(.....)).....)))))...(((..(((((.........))).)).)))....(((((((((((....))))))))))).. ( -24.80) >DroYak_CAF1 65304 97 - 1 A-------AAAAUGUUCCUAAG-----UCUAAGGCAAAUCAACCUCUUGACUUCCCAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCU .-------..............-----.....(((...........(((((..(((((.........))).)).)))))...((((((((((....))))))))))))) ( -24.40) >DroMoj_CAF1 94751 104 - 1 -----AAAAGACGAAUCCGAAAACAAAACAAAAACAAACAAAAUUAUUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAUGCU -----.......................................(((((((..(((((..........))))).)))))))..(((((((((....))))))))).... ( -23.20) >consensus A_______AAAAGGUUCCUAAG_____UCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCU ................................(((...........(((((..(((..............))).)))))...((((((((((....))))))))))))) (-16.96 = -17.99 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:57 2006