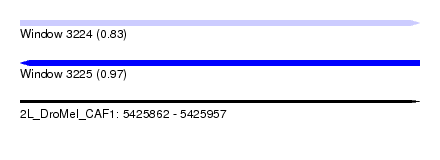
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,425,862 – 5,425,957 |
| Length | 95 |
| Max. P | 0.971586 |

| Location | 5,425,862 – 5,425,957 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -15.46 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5425862 95 + 22407834 GCCACCAGCCGCCAUUGUUUUUCAACAUUUUUAUGGUAAAAUGUUAAAUGAGCUGGGCGGCACUCGCACUCGCUGUUGGAUGAUGGGGAUUUUAU .((.(((((((((...((((...((((((((......))))))))....))))..)))))).((.(((.....))).))....)))))....... ( -29.30) >DroSec_CAF1 45569 72 + 1 GCCACCAGCCGCCAUUGUUUUUCAACAUUUUUAUGGUAAAAUGUUAAAUGAGCUGGG-----------------------UGGUGGGGAUUUUAU .((.(((.(((((...((((...((((((((......))))))))....))))..))-----------------------))))))))....... ( -23.30) >DroSim_CAF1 56603 72 + 1 GCCACCAGCCGCCAUUGUUUUUCAACAUUUUUAUGGUAAAAUGUUAAAUGAGCUGGG-----------------------UGGUGGGGAUUUUAU .((.(((.(((((...((((...((((((((......))))))))....))))..))-----------------------))))))))....... ( -23.30) >DroEre_CAF1 44435 85 + 1 GCCACCCGCCGCCAUUGUUUUUCAACAUUUUUAUGGUAAAAUGUUAAAUGAGCUGGGCGGCAAUU------GCUGU----UGGUGGGGAUUUUAU .(((((.((((((...((((...((((((((......))))))))....))))..))))))(((.------...))----))))))......... ( -29.30) >DroYak_CAF1 52071 85 + 1 GCCACCAGCCGCCAUUGUUUUUCAACAUUUUUAUGGUAAAAUGUUAAAUGAGCUGGGCGGCAAUC------GCUGG----UGGUGGGGAUUUUAU (((((((((((((...((((...((((((((......))))))))....))))..))))))....------..)))----))))........... ( -31.50) >DroAna_CAF1 48205 82 + 1 UCAUAGCGACGCCAUUGUUUUUCAACAUUUUUAUGGUAAAAUGUUAAAUGAGCUGGGCGGCG---------GCCGA----CAAGGGGGUUUUUAU ..((((.(((.((.(((((....((((((((......))))))))..........(((....---------)))))----))).)).))).)))) ( -19.60) >consensus GCCACCAGCCGCCAUUGUUUUUCAACAUUUUUAUGGUAAAAUGUUAAAUGAGCUGGGCGGCA_________GCUG_____UGGUGGGGAUUUUAU .(((((....(((...((((...((((((((......))))))))....))))..)))(((..........))).......)))))......... (-15.46 = -16.35 + 0.89)


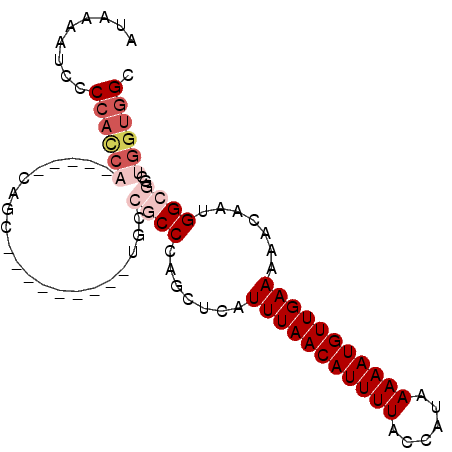
| Location | 5,425,862 – 5,425,957 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -13.10 |
| Energy contribution | -14.66 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.33 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5425862 95 - 22407834 AUAAAAUCCCCAUCAUCCAACAGCGAGUGCGAGUGCCGCCCAGCUCAUUUAACAUUUUACCAUAAAAAUGUUGAAAAACAAUGGCGGCUGGUGGC .........((((((.......((....))....((((((.......(((((((((((......))))))))))).......)))))))))))). ( -28.24) >DroSec_CAF1 45569 72 - 1 AUAAAAUCCCCACCA-----------------------CCCAGCUCAUUUAACAUUUUACCAUAAAAAUGUUGAAAAACAAUGGCGGCUGGUGGC .........((((((-----------------------.((.((.(((((((((((((......))))))))))......))))))).)))))). ( -21.60) >DroSim_CAF1 56603 72 - 1 AUAAAAUCCCCACCA-----------------------CCCAGCUCAUUUAACAUUUUACCAUAAAAAUGUUGAAAAACAAUGGCGGCUGGUGGC .........((((((-----------------------.((.((.(((((((((((((......))))))))))......))))))).)))))). ( -21.60) >DroEre_CAF1 44435 85 - 1 AUAAAAUCCCCACCA----ACAGC------AAUUGCCGCCCAGCUCAUUUAACAUUUUACCAUAAAAAUGUUGAAAAACAAUGGCGGCGGGUGGC .........(((((.----.....------...(((((((.......(((((((((((......))))))))))).......)))))))))))). ( -27.24) >DroYak_CAF1 52071 85 - 1 AUAAAAUCCCCACCA----CCAGC------GAUUGCCGCCCAGCUCAUUUAACAUUUUACCAUAAAAAUGUUGAAAAACAAUGGCGGCUGGUGGC ............(((----(((..------....((((((.......(((((((((((......))))))))))).......)))))))))))). ( -28.54) >DroAna_CAF1 48205 82 - 1 AUAAAAACCCCCUUG----UCGGC---------CGCCGCCCAGCUCAUUUAACAUUUUACCAUAAAAAUGUUGAAAAACAAUGGCGUCGCUAUGA ...............----..(((---------.(.((((.......(((((((((((......))))))))))).......)))).)))).... ( -17.04) >consensus AUAAAAUCCCCACCA_____CAGC_________UGCCGCCCAGCUCAUUUAACAUUUUACCAUAAAAAUGUUGAAAAACAAUGGCGGCUGGUGGC .........((((((.....................((((.......(((((((((((......))))))))))).......))))..)))))). (-13.10 = -14.66 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:53 2006