
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,422,565 – 5,422,673 |
| Length | 108 |
| Max. P | 0.598951 |

| Location | 5,422,565 – 5,422,673 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5422565 108 - 22407834 CAAGUGGCUAAAAGUGUCAAUUCGUGAAAAU-UUCACAAAUUCCAAUCGAUGUGAAUAAUCAGAAUUGGCGUCGACAACAACAGCAGCAGCAGCACCAGCACAAAAGCC ...((((((.....((((.....((((....-.))))...........(((((.(((.......))).))))))))).....))).((....)).....)))....... ( -22.90) >DroSec_CAF1 42375 99 - 1 CAAGUGGCUAAAAGUGUCAAUUCGUGAAAAU-UUCACAAAUUCCAAUCGAUGUGAAUAAUCAGAAUUGGCGGCGACAACAACGGCAGCAGCA---------CAAAAGCC .....((((....((((((((((.(((....-((((((............))))))...)))))))))(((.((.......)).).)).)))---------)...)))) ( -22.40) >DroEre_CAF1 40820 96 - 1 CAAGUGGCUAAAAGUGUCAAUUCGUGAAAAU-UUCACAAAUUCCAAUCGAUGUGAAUAAUCAGAAUUGGCGGCGACAACAA---------CAG---CAGCACAAAAGCC .....((((.....(((((((((.(((....-((((((............))))))...))))))))))))((..(.....---------..)---..)).....)))) ( -21.80) >DroWil_CAF1 61532 95 - 1 CAAGUGGCUAGAAGUGUCAAUUCGUGAAAGUUUUCACAAAUUCCAAUCGAUGUGAAUAAUCAGAAACGGAAUCGAUAGAAACG-CAACA-------------AAGAACA ...((.((......((((.....(((((....)))))..(((((..(((((.......))).))...))))).)))).....)-).)).-------------....... ( -13.70) >DroYak_CAF1 48596 105 - 1 CAAGUGGCUAGGAGUGUCAAUUCGUGAAAAU-UUCACAAAUUCCAAUCGAUGUGAAUAAUCAGAAUUGGCGGCGACAACAACAGCAACAACAG---CAGCACAAAAGCC .....((((.(..((((((((((.(((....-((((((............))))))...))))))))))).))..).......((.......)---)........)))) ( -23.40) >DroAna_CAF1 45103 94 - 1 CAAGUGGCGGCUAGUGUCAAUUCGUGAAAAU-UUCACAAAUUCCAAUCGAUGUGAAUAAUCAGAAUUGGCGGCGACAAUAACAAUUG--------------GGAUUGCU .....((((((((((((((((((.(((....-((((((............))))))...)))))))))))(....).......))))--------------)..))))) ( -20.90) >consensus CAAGUGGCUAAAAGUGUCAAUUCGUGAAAAU_UUCACAAAUUCCAAUCGAUGUGAAUAAUCAGAAUUGGCGGCGACAACAACAGCAGCA_CA_________CAAAAGCC .....((((......(((((((((((((....)))))..((((((.....)).)))).....))))))))(....).............................)))) (-15.93 = -16.10 + 0.17)

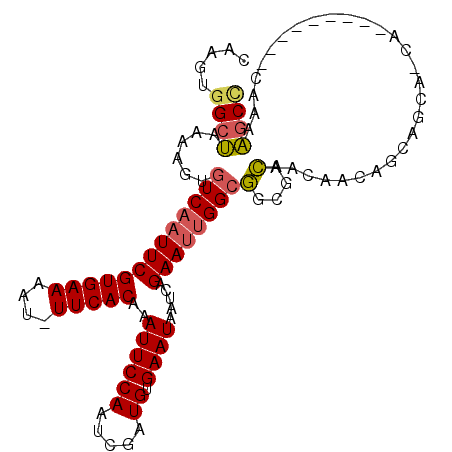
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:50 2006