| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,872 – 12,982 |
| Length | 110 |
| Max. P | 0.980333 |

| Location | 12,872 – 12,982 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -17.27 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
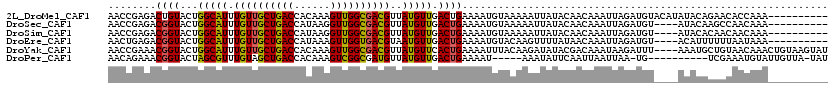
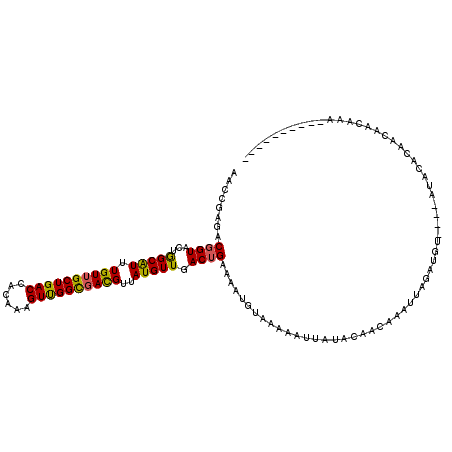
>2L_DroMel_CAF1 12872 110 + 22407834 AACCGAGACUGUACUGGCAUUUGUUGCUGACCACAAAGUUGGCGACGUUAUGUUGACUGAAAAUGUAAAAAUUAUACAACAAAUUAGAUGUACAUAUACAGAACACCAAA---------- ........(((((...(((((((((((..((......))..)))))....(((((.....................)))))....)))))).....))))).........---------- ( -22.10) >DroSec_CAF1 1102 106 + 1 AACCGAGACGGUACUGGCAUUUGUUGCUGACCAUAAGGUUGGCGACGUUAUGUUGACUGAAAAUGUAAAAAUUAUACAACAAAUUAGAUGU----AUACAAGCCAACAAA---------- .((((...))))..((((....(((((..(((....)))..))))).(((((((.......)))))))....((((((.(......).)))----)))...)))).....---------- ( -28.50) >DroSim_CAF1 1102 106 + 1 AACCGAGACGGUACUGGCAUUUGUUGCUGACCAUAAGGUUGGCGACGUUAUGUUGACUGAAAAUGUAAAAAUUAUACAACAAAUUAGAUGU----AUACACAACAACAAA---------- ........((((...(((((.((((((..(((....)))..))))))..))))).))))....(((......((((((.(......).)))----))).......)))..---------- ( -25.62) >DroEre_CAF1 1435 106 + 1 AACUGAGACGGUACUGGCAUUUGUUGCUGACCAUAAAGUUGGUGACGUAAUGUUGACUGAAAAUGUACAAGUUUUAUAACAAAUUAGAUGU----ACAUUUUUUAAUAAA---------- ........((((...(((((((((..(..((......))..)..))).)))))).))))((((((((((...((((........)))))))----)))))))........---------- ( -25.90) >DroYak_CAF1 1092 116 + 1 AACCGAAACGGUACUGGCAUUUGUUGCUGACCACAAAGUUGGCGACGUUAUGUUCACUGAAAAUUUACAAGAUAUACGACAAAUAAGAUUU----AAAUGCUGUAACAAACUGUAAGUAU .......(((((((.((((((((((((..((......))..)))))((..(((...((...........))..)))..))...........----))))))))).....)))))...... ( -22.30) >DroPer_CAF1 7891 103 + 1 AACAGAAACGGUACUAGCGUUUGUAGCUGACCACAAAGUCGGCGAUGUUAUGUUGACUGAAAAU-----AAAUAUUCAAUUAAUUAA-UG----------UCGAAAUGUAUUGUUA-UAU ......((((((((...........((((((......))))))(((((((.(((((.((((...-----.....)))).))))))))-))----------)).....)))))))).-... ( -24.90) >consensus AACCGAGACGGUACUGGCAUUUGUUGCUGACCACAAAGUUGGCGACGUUAUGUUGACUGAAAAUGUAAAAAUUAUACAACAAAUUAGAUGU____AUACACAACAACAAA__________ ........((((...(((((.((((((((((......))))))))))..))))).))))............................................................. (-17.27 = -16.92 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:22:57 2006