
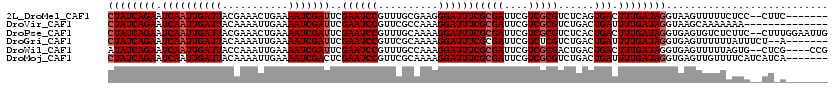
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,414,116 – 5,414,227 |
| Length | 111 |
| Max. P | 0.615998 |

| Location | 5,414,116 – 5,414,227 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -20.93 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
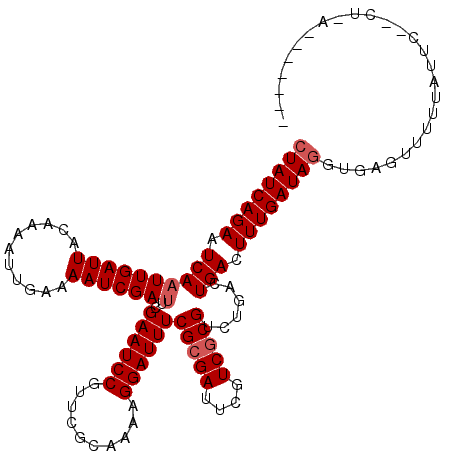
>2L_DroMel_CAF1 5414116 111 + 22407834 CUAUCAGAAUCAAUUGAUUACGAAACUGAAAAUCGAUUCGAAUCCGUUUGCGAAGGGAUUUCGCGAUUCGUCGCGUCUCAGUGACUUUGAUAGGUAAGUUUUUCUCC--CUUC------- ((((((((((((((.((((.((((.(........).)))))))).))).((((((...)))))))))))(((((......)))))..))))))..............--....------- ( -28.80) >DroVir_CAF1 50898 106 + 1 CUAUCAGAAUCAAUUGAUUACAAAAUUGAAAAUCGAUUCGAAUCCGUUCGCCAAAGGAUUUCGCGAUUCGUCGCGUCUGACUGAUUUUGAUAGGUAAGCAAAAAAA-------------- (((((((((((((((((((...........)))))))((((((((..........))))))(((((....)))))...)).)))))))))))).............-------------- ( -27.10) >DroPse_CAF1 40820 118 + 1 CUAUCAGAAUCAAUUGAUUACGAAACUGAAAAUCGAUUCGAAUCCGUUUGCAAAAGGAUUUCGCGAUUCGUCGCGUCUCACUGACUUUGAUAGGUGAGUGUCUCUUC--CUUUGGAAUUG .........((((((((((...........)))))))).)).((((....).((((((...(((((....)))))...((((.((((....)))).)))).....))--))))))).... ( -26.00) >DroGri_CAF1 56076 111 + 1 CUAUCAGAAUCAAUUGAUUACAAAAUUGAAAAUCGAUUCGAAUCCGUUCGCAAAAGGAUUUCGCGAUUCGUCUCGUCUGACUGAUUUUGAUAGGUGAGUUUUUUAUUUCU--A------- ((((((((((((.(..(((....)))..)....(((..((((((((.((.......))...)).))))))..)))......)))))))))))).................--.------- ( -26.80) >DroWil_CAF1 49143 114 + 1 AUAUCAGAAUCAAUUGAUUACCAAAUUGAAAAUCGAUUCGAAUCCGUUUGCCAAAGGAUUUCGCGAUUCGUCGCGACUGACUGACUUUGAUAGGUGAGUUUUUAGUG--CUCG----CCG .(((((((.((((((........))))))...(((...((((((((....((...))....)).))))))...))).........)))))))(((((((.......)--))))----)). ( -33.10) >DroMoj_CAF1 57136 113 + 1 CUAUCAGAAUCAAUUGAUUACAAAAUUGAAAAUCGACUCGAAUCCGUUCGCAAAAGGAUUUCGCGAUUCGUCGCGUCUGACUGAUUUUGAUAGGUGAGUUGUUUUCAUCAUCA------- .(((((((((((.(..(((....)))..).....(((..((((((..........)))))).((((....)))))))....)))))))))))((((((.....))))))....------- ( -30.10) >consensus CUAUCAGAAUCAAUUGAUUACAAAAUUGAAAAUCGAUUCGAAUCCGUUCGCAAAAGGAUUUCGCGAUUCGUCGCGUCUGACUGACUUUGAUAGGUGAGUUUUUAUUC__CU_A_______ ((((((((.((((((((((...........)))))))..((((((..........))))))(((((....)))))......))).))))))))........................... (-20.93 = -21.43 + 0.50)
| Location | 5,414,116 – 5,414,227 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5414116 111 - 22407834 -------GAAG--GGAGAAAAACUUACCUAUCAAAGUCACUGAGACGCGACGAAUCGCGAAAUCCCUUCGCAAACGGAUUCGAAUCGAUUUUCAGUUUCGUAAUCAAUUGAUUCUGAUAG -------....--..............((((((.(((((((((((..((((((((((((((.....))))).....))))))..)))..)))))))...(....)....)))).)))))) ( -26.90) >DroVir_CAF1 50898 106 - 1 --------------UUUUUUUGCUUACCUAUCAAAAUCAGUCAGACGCGACGAAUCGCGAAAUCCUUUGGCGAACGGAUUCGAAUCGAUUUUCAAUUUUGUAAUCAAUUGAUUCUGAUAG --------------.............((((((.((((((((((((((((....)))))......))))))(((((((...(((......)))...)))))..))...))))).)))))) ( -23.10) >DroPse_CAF1 40820 118 - 1 CAAUUCCAAAG--GAAGAGACACUCACCUAUCAAAGUCAGUGAGACGCGACGAAUCGCGAAAUCCUUUUGCAAACGGAUUCGAAUCGAUUUUCAGUUUCGUAAUCAAUUGAUUCUGAUAG ...((((...)--)))(((...)))..((((((.(((((((((.(((.(((((((((((((.(((..........)))))))...))))))...))).)))..)).))))))).)))))) ( -27.60) >DroGri_CAF1 56076 111 - 1 -------U--AGAAAUAAAAAACUCACCUAUCAAAAUCAGUCAGACGAGACGAAUCGCGAAAUCCUUUUGCGAACGGAUUCGAAUCGAUUUUCAAUUUUGUAAUCAAUUGAUUCUGAUAG -------.--.................((((((.(((((((...((((((((..(((((((.....))))))).)).....(((......)))..)))))).....))))))).)))))) ( -23.50) >DroWil_CAF1 49143 114 - 1 CGG----CGAG--CACUAAAAACUCACCUAUCAAAGUCAGUCAGUCGCGACGAAUCGCGAAAUCCUUUGGCAAACGGAUUCGAAUCGAUUUUCAAUUUGGUAAUCAAUUGAUUCUGAUAU .((----.(((--.........))).))(((((.((((((((((((((((....))))))......)))))...((((((.(((......))))))))).........))))).))))). ( -27.20) >DroMoj_CAF1 57136 113 - 1 -------UGAUGAUGAAAACAACUCACCUAUCAAAAUCAGUCAGACGCGACGAAUCGCGAAAUCCUUUUGCGAACGGAUUCGAGUCGAUUUUCAAUUUUGUAAUCAAUUGAUUCUGAUAG -------....................((((((.(((((((..(((.(((((..(((((((.....))))))).))...))).)))((((..((....)).)))).))))))).)))))) ( -26.00) >consensus _______U_AG__GAAAAAAAACUCACCUAUCAAAAUCAGUCAGACGCGACGAAUCGCGAAAUCCUUUUGCAAACGGAUUCGAAUCGAUUUUCAAUUUUGUAAUCAAUUGAUUCUGAUAG ...........................((((((.(((((((....(((((....)))))..............(((((...(((......)))...))))).....))))))).)))))) (-19.54 = -19.65 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:47 2006