| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,411,755 – 5,411,856 |
| Length | 101 |
| Max. P | 0.996912 |

| Location | 5,411,755 – 5,411,856 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.96 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -13.13 |
| Energy contribution | -15.17 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
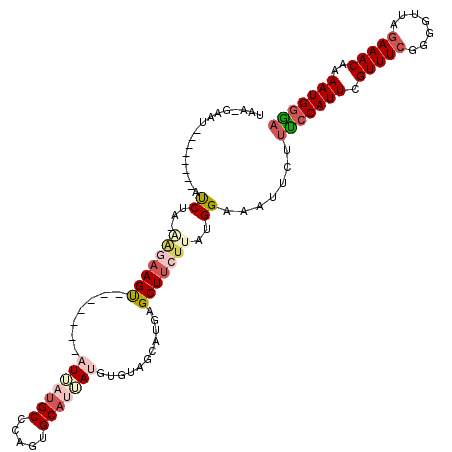
>2L_DroMel_CAF1 5411755 101 + 22407834 UAA-GAAU--------ACCUU-GAGAAGU-------AUUAUGCCCAGUGCAUUAUGUGUAGCAUGAGCUUCUUAUGGAAAUUCUUUCCAUUCGUUUCGGGGUUAGAAACAAAAUGGGA .((-((((--------.((.(-(((((((-------.((((((....(((((...))))))))))))))))))).))..))))))((((((.(((((.......)))))..)))))). ( -29.90) >DroSec_CAF1 31723 101 + 1 UAA-GAAA--------AUCUA-AAGAAGU-------AUUAUGCCCAGUGCAUUAUGUGUAGCAUGAGCUUCUUAUGGAAAUUCUUUCCAUUCGUUUCGGAGUUAGAAACAAAAUGGGA .((-(((.--------.((((-(((((((-------.((((((....(((((...)))))))))))))))))).))))..)))))((((((.(((((.......)))))..)))))). ( -28.20) >DroSim_CAF1 31084 101 + 1 UAA-AAAU--------AUCUA-AGGAAGU-------AUAAUGCACAGUGCAUUAUGUAUAGCAUGAGCUUCUUAUGGAAAUUCUUCCCAUUCGUUUCGGGGUUAGAAACAAAAUGGGA ...-....--------.((((-(((((((-------...((((...((((.....)))).))))..))))))).))))......(((((((.(((((.......)))))..))))))) ( -27.90) >DroEre_CAF1 29941 112 + 1 UAU-UUUUUCAUUGAGUUCUU-----AGCCUGGUUGUUUUUGCCCGGUGCAUGAUGUGUAGAAUGCGCUUCUUAUGGAAAUUCUCUCCAUUCGUUUCGGGGGUAAAAACAAAAUGGAA ...-.................-----..((...(((((((((((((((((((..........)))))))....(((((.......))))).........))))))))))))...)).. ( -30.40) >DroYak_CAF1 37527 118 + 1 AAUAUUUUUCAUUGAGUUCUCACAUAAGCCUUGUGCUUUUCGCCCAGUGCAUGAUGUGUAGAAAGAGCUUCUUAUGGAAAUUCUUCACAUUCGUUUCGGGGGUAGAAACAAAAUGGGA .....(((((((.((((((((((((..((.(((.((.....)).))).))...))))).....)))))))...)))))))...(((.((((.(((((.......)))))..))))))) ( -26.10) >consensus UAA_GAAU________AUCUA_AAGAAGU_______AUUAUGCCCAGUGCAUUAUGUGUAGCAUGAGCUUCUUAUGGAAAUUCUUUCCAUUCGUUUCGGGGUUAGAAACAAAAUGGGA .................((...(((((((.......(((((((.....)))))))...........)))))))..)).......(((((((.(((((.......)))))..))))))) (-13.13 = -15.17 + 2.04)


| Location | 5,411,755 – 5,411,856 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.96 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.96 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
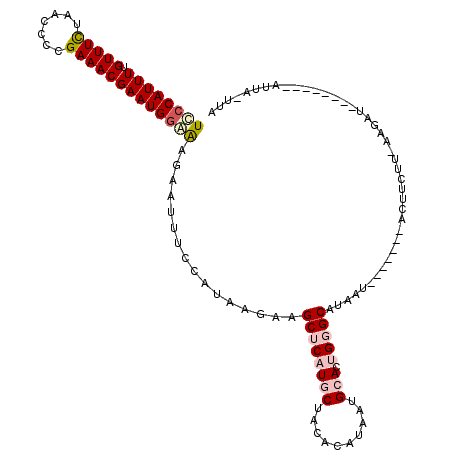
>2L_DroMel_CAF1 5411755 101 - 22407834 UCCCAUUUUGUUUCUAACCCCGAAACGAAUGGAAAGAAUUUCCAUAAGAAGCUCAUGCUACACAUAAUGCACUGGGCAUAAU-------ACUUCUC-AAGGU--------AUUC-UUA ..((((((.(((((.......))))))))))).((((((..((...((((((((((((..........))).))))).....-------..)))).-..)).--------))))-)). ( -23.81) >DroSec_CAF1 31723 101 - 1 UCCCAUUUUGUUUCUAACUCCGAAACGAAUGGAAAGAAUUUCCAUAAGAAGCUCAUGCUACACAUAAUGCACUGGGCAUAAU-------ACUUCUU-UAGAU--------UUUC-UUA ..((((((.(((((.......))))))))))).(((((..((...(((((((((((((..........))).))))).....-------..)))))-..)).--------.)))-)). ( -24.51) >DroSim_CAF1 31084 101 - 1 UCCCAUUUUGUUUCUAACCCCGAAACGAAUGGGAAGAAUUUCCAUAAGAAGCUCAUGCUAUACAUAAUGCACUGUGCAUUAU-------ACUUCCU-UAGAU--------AUUU-UUA .(((((((.(((((.......))))))))))))((((((.((..((((.(((....)))....(((((((.....)))))))-------.....))-)))).--------))))-)). ( -25.80) >DroEre_CAF1 29941 112 - 1 UUCCAUUUUGUUUUUACCCCCGAAACGAAUGGAGAGAAUUUCCAUAAGAAGCGCAUUCUACACAUCAUGCACCGGGCAAAAACAACCAGGCU-----AAGAACUCAAUGAAAAA-AUA ...((((..(((((((((((((...(..(((((((...)))))))..)....((((..........))))..))))............)).)-----))))))..)))).....-... ( -21.30) >DroYak_CAF1 37527 118 - 1 UCCCAUUUUGUUUCUACCCCCGAAACGAAUGUGAAGAAUUUCCAUAAGAAGCUCUUUCUACACAUCAUGCACUGGGCGAAAAGCACAAGGCUUAUGUGAGAACUCAAUGAAAAAUAUU ...(((((((((((.......)))))))).)))((((.((((.....)))).))))(((.(((((...((.((..((.....))...))))..))))))))................. ( -23.20) >consensus UCCCAUUUUGUUUCUAACCCCGAAACGAAUGGAAAGAAUUUCCAUAAGAAGCUCAUGCUACACAUAAUGCACUGGGCAUAAU_______ACUUCUU_AAGAU________AUUA_UUA ((((((((.(((((.......)))))))))))))................((((((((..........))).)))))......................................... (-12.64 = -13.96 + 1.32)

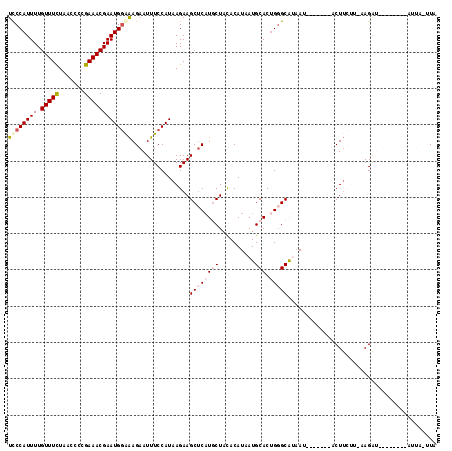
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:44 2006