
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,390,209 – 5,390,302 |
| Length | 93 |
| Max. P | 0.943705 |

| Location | 5,390,209 – 5,390,302 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.71 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
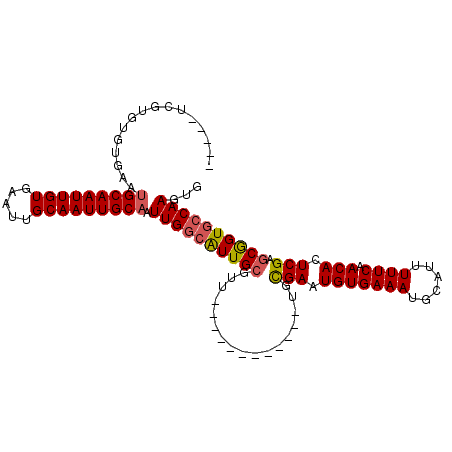
>2L_DroMel_CAF1 5390209 93 + 22407834 CACUUGGCACCGUUCGAGUGUUGAAAAAUGCAUUUCACAUUCACA------------AACGCAAUGCCAAUUGCAAUUGCAAUUCACAAUUGCAUUCACACACGA----- ...((((((.((((.((((((.((((......))))))))))...------------))))...)))))).((((((((.......))))))))...........----- ( -24.90) >DroSec_CAF1 10663 93 + 1 CACUUGGCACCGCUCGAGUGUUGAAAAAUGAAUUUCACAUUCGCA------------AACGCAAUGCCAAUUGCAAUUGCAAUUCACAAUUGCAUUCACACACGA----- ...((((((..((.(((((((.((((......)))))))))))..------------...))..)))))).((((((((.......))))))))...........----- ( -25.90) >DroSim_CAF1 9648 93 + 1 CACUUGGCACCGCUCGAGUGUUGAAAAAUGCAUUUCACAUUCGCA------------AACGCAAUGCCAAUUGCAAUUGCAAUUCACAAUUGCAUUCACACACGA----- ...((((((..((.(((((((.((((......)))))))))))..------------...))..)))))).((((((((.......))))))))...........----- ( -25.90) >DroEre_CAF1 8071 102 + 1 CACUUGGCACCGCUCGAGUGUUGAAAAAUGCAUUUCACAUUCGCA-ACGCAAAGGCA-ACGCAACGCCAAUUGCAAUUGC-AUUCACAAUUGCAUUCACACACGA----- ...(((((...((.(((((((.((((......)))))))))))..-..))...(...-.).....))))).((((((((.-.....))))))))...........----- ( -27.70) >DroYak_CAF1 15249 105 + 1 CACUUGGCACCGCUCGAGUGUUGAAAAAUGCAUUUCACAUUCGCAAACGAAAACGCAAACGCAACGCCAAUUGCAAUUGCUAUUCACAAUUGCAUUCACACACGA----- ...(((((..((..(((((((.((((......)))))))))))....)).....((....))...))))).((((((((.......))))))))...........----- ( -26.00) >DroAna_CAF1 11560 98 + 1 CAGUUGGCACUGCUCGAGUGUUGAAAAAUGCAUUUCACAUUCGCA------------AUUGCAAUCGCAAUGGCAAUUGCAGCUUACAAUUGCUUUCACACACGAGUGGC .........(..((((.((((.((((................(((------------(((((..........)))))))).((........)))))))))).))))..). ( -28.60) >consensus CACUUGGCACCGCUCGAGUGUUGAAAAAUGCAUUUCACAUUCGCA____________AACGCAAUGCCAAUUGCAAUUGCAAUUCACAAUUGCAUUCACACACGA_____ ...(((((.......((((((.((((......))))))))))((................))...))))).((((((((.......))))))))................ (-20.48 = -20.71 + 0.22)



| Location | 5,390,209 – 5,390,302 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -27.03 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5390209 93 - 22407834 -----UCGUGUGUGAAUGCAAUUGUGAAUUGCAAUUGCAAUUGGCAUUGCGUU------------UGUGAAUGUGAAAUGCAUUUUUCAACACUCGAACGGUGCCAAGUG -----...........(((((((((.....))))))))).(((((((..((((------------((.(....(((((......)))))...).)))))))))))))... ( -29.50) >DroSec_CAF1 10663 93 - 1 -----UCGUGUGUGAAUGCAAUUGUGAAUUGCAAUUGCAAUUGGCAUUGCGUU------------UGCGAAUGUGAAAUUCAUUUUUCAACACUCGAGCGGUGCCAAGUG -----...........(((((((((.....))))))))).((((((((((...------------..(((.(((((((......)))).))).))).))))))))))... ( -33.70) >DroSim_CAF1 9648 93 - 1 -----UCGUGUGUGAAUGCAAUUGUGAAUUGCAAUUGCAAUUGGCAUUGCGUU------------UGCGAAUGUGAAAUGCAUUUUUCAACACUCGAGCGGUGCCAAGUG -----...........(((((((((.....))))))))).((((((((((...------------..(((.(((((((......)))).))).))).))))))))))... ( -33.70) >DroEre_CAF1 8071 102 - 1 -----UCGUGUGUGAAUGCAAUUGUGAAU-GCAAUUGCAAUUGGCGUUGCGU-UGCCUUUGCGU-UGCGAAUGUGAAAUGCAUUUUUCAACACUCGAGCGGUGCCAAGUG -----...........(((((((((....-))))))))).((((((..((..-.((....))((-((.((((((.....))))))..))))......))..))))))... ( -32.30) >DroYak_CAF1 15249 105 - 1 -----UCGUGUGUGAAUGCAAUUGUGAAUAGCAAUUGCAAUUGGCGUUGCGUUUGCGUUUUCGUUUGCGAAUGUGAAAUGCAUUUUUCAACACUCGAGCGGUGCCAAGUG -----(((.((((((((((((((((.....)))))))))....((((((((((((((........)))))))))..)))))....))).)))).)))............. ( -34.30) >DroAna_CAF1 11560 98 - 1 GCCACUCGUGUGUGAAAGCAAUUGUAAGCUGCAAUUGCCAUUGCGAUUGCAAU------------UGCGAAUGUGAAAUGCAUUUUUCAACACUCGAGCAGUGCCAACUG ....((((.(((((((.((((((((((..((((((....)))))).)))))))------------)))((((((.....))))))))).)))).))))((((....)))) ( -35.50) >consensus _____UCGUGUGUGAAUGCAAUUGUGAAUUGCAAUUGCAAUUGGCAUUGCGUU____________UGCGAAUGUGAAAUGCAUUUUUCAACACUCGAGCGGUGCCAAGUG ................(((((((((.....))))))))).((((((((((.................(((.(((((((......)))).))).))).))))))))))... (-27.03 = -27.20 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:38 2006