
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 595,416 – 595,563 |
| Length | 147 |
| Max. P | 0.914638 |

| Location | 595,416 – 595,526 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
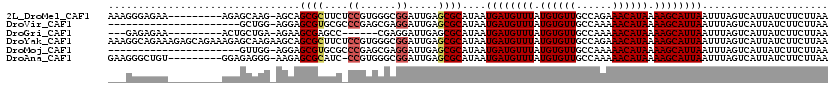
>2L_DroMel_CAF1 595416 110 - 22407834 AAAGGGAGAA---------AGAGCAAG-AGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGCCAGAAACAUAAAAGCAUUAAUUUAGUCAUUAUCUUCUUAA ..(((((((.---------...((...-.)).((((((.((((....))))..))))))....((((((((.((((((......)))))).))))))))............))))))).. ( -34.80) >DroVir_CAF1 33154 97 - 1 ----------------------GCUGG-AGGAGCGUGCGCCCGAGCGAGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGCCAAAAACAUAAAAGCAUUAAUUUAGUCAUUAUCUUCUUAA ----------------------...((-((((..((((((.(((.......))).))))))..((((((((.((((((......)))))).))))))))............))))))... ( -28.20) >DroGri_CAF1 32486 101 - 1 ---GAGAGAA---------ACUGCUGA-AGAAGCGAGCC------CGAGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGCCAAAAACAUAAAAGCAUUAAUUUAGUCAUUAUCUUCUUAA ---(((((((---------(..(((((-(...(((..(.------........)..)))....((((((((.((((((......)))))).)))))))).))))))..)).)).)))).. ( -21.70) >DroYak_CAF1 27025 120 - 1 AAAGGCAGAAAGAGCAGAAAGAGCAAGAAGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGCCAGAAACAUAAAAGCAUUAAUUUAGUCAUUAUCUUCUUAA .............((.......))((((((..((((((.((((....))))..))))))....((((((((.((((((......)))))).)))))))).............)))))).. ( -33.20) >DroMoj_CAF1 31149 97 - 1 ----------------------GUUGG-AGGAGCGUGCGCCCGAGCGAGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGCCAAAAACAUAAAAGCAUUAAUUUAGUCAUUAUCUUCUUAA ----------------------...((-((((..((((((.(((.......))).))))))..((((((((.((((((......)))))).))))))))............))))))... ( -28.20) >DroAna_CAF1 27377 109 - 1 GAAGGGCUGU---------GGAGAGGG-AAGAGCGCAUC-CCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGCCAAAAACAUAAAAGCAUUAAUUUAGUCAUUAUCUUCUUAA (((((...((---------((..(((.-....((((.((-(((....)))...))))))....((((((((.((((((......)))))).)))))))).)))..))))..))))).... ( -30.70) >consensus ___GGGAGAA_________AGAGCUGG_AGGAGCGCGCC_CCGUGCGAGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGCCAAAAACAUAAAAGCAUUAAUUUAGUCAUUAUCUUCUUAA ................................((((....((......)).....))))....((((((((.((((((......)))))).))))))))..................... (-18.10 = -18.43 + 0.33)

| Location | 595,456 – 595,563 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -19.61 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
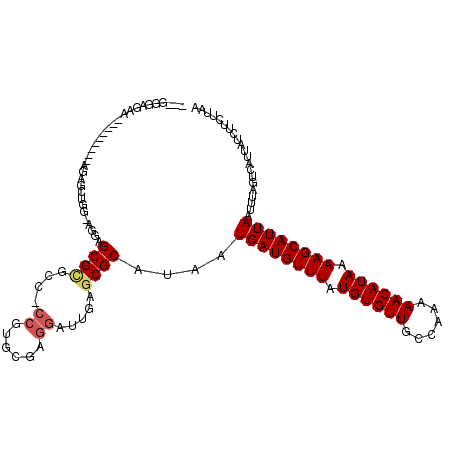
>2L_DroMel_CAF1 595456 107 + 22407834 GCAACACAUAAACAUCAUUAUGCGCUCAAUCCGCCCACGGAGAAGCGCUGCU-CUUGCUCU---------UUCUCCCUUUCUGAGCUUUAUCUUUAUCAU---CAUUCUUGCAGACGUUG ((((.................(((((...((((....))))..)))))....-...((((.---------............))))..............---.....))))........ ( -21.42) >DroSec_CAF1 26778 110 + 1 GCAACACAUAAACAUCAUUAUGCGCUCAAUCCGCCCACGGAGAAGCGCUGCU-CUUGCUCU---------UUCUCCCUUUUCCAGCUUUAUCUUUAUCAUCUCCAUUCUUGCAGACGUUG .((((..((((((........(((((...((((....))))..))))).((.-...))...---------..............).)))))........(((.((....)).))).)))) ( -18.40) >DroSim_CAF1 25346 110 + 1 GCAACACAUAAACAUCAUUAUGCGCUCAAUCCGCCCACGGAGAAGCGCUGCU-CUUGCUCU---------UUCUCCCUUUGCCAGCUUUAUCUUUAUCAUCUCCAUUCUUGCAGACGUUG .((((................(((((...((((....))))..))))).(((-...((...---------..........)).))).............(((.((....)).))).)))) ( -19.12) >DroEre_CAF1 27123 111 + 1 GCAACACAUAAACAUCAUUAUGCGCUCAAUCCGCCCACGGAGAAGCGCUGCUUCUUGCUCU---------UUCUGCCUUUUCCAGCUUUAUCUUUAUCAUCUCCAUUCUUGCAGAUGUUA ..........((((((....((((((...((((....))))..)))((((......((...---------....))......))))........................))))))))). ( -22.10) >DroYak_CAF1 27065 109 + 1 GCAACACAUAAACAUCAUUAUGCGCUCAAUCCGCCCACGGAGAAGCGCUGCUUCUUGCUCUUUCUGCUCUUUCUGCCUUUUCCAGCUUUAUCUUUAUCAUCUCCGUUCU----------- (((..................(((((...((((....))))..))))).((.....))......)))..........................................----------- ( -17.00) >consensus GCAACACAUAAACAUCAUUAUGCGCUCAAUCCGCCCACGGAGAAGCGCUGCU_CUUGCUCU_________UUCUCCCUUUUCCAGCUUUAUCUUUAUCAUCUCCAUUCUUGCAGACGUUG ((...................(((((...((((....))))..))))).((.....))..........................)).................................. (-16.40 = -16.40 + 0.00)


| Location | 595,456 – 595,563 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -21.44 |
| Energy contribution | -23.12 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
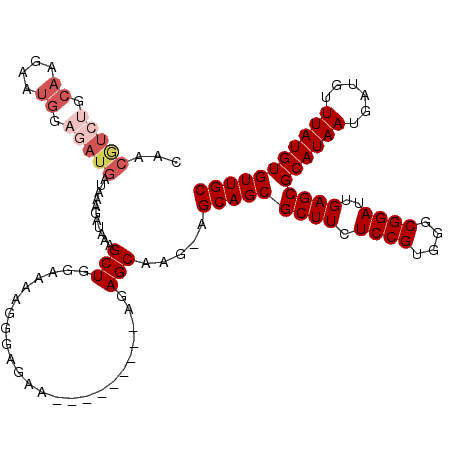
>2L_DroMel_CAF1 595456 107 - 22407834 CAACGUCUGCAAGAAUG---AUGAUAAAGAUAAAGCUCAGAAAGGGAGAA---------AGAGCAAG-AGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGC ...((((..(.......---........((......))......((((((---------.(.((...-....)).))))))))..)))).....(((((((((......))))))))).. ( -28.10) >DroSec_CAF1 26778 110 - 1 CAACGUCUGCAAGAAUGGAGAUGAUAAAGAUAAAGCUGGAAAAGGGAGAA---------AGAGCAAG-AGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGC ...((((..(.........................((.....))((((((---------.(.((...-....)).))))))))..)))).....(((((((((......))))))))).. ( -28.30) >DroSim_CAF1 25346 110 - 1 CAACGUCUGCAAGAAUGGAGAUGAUAAAGAUAAAGCUGGCAAAGGGAGAA---------AGAGCAAG-AGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGC ...(((((.((....)).))))).....(((...(((.((....((((((---------.(.((...-....)).))))))))).)))..))).(((((((((......))))))))).. ( -30.80) >DroEre_CAF1 27123 111 - 1 UAACAUCUGCAAGAAUGGAGAUGAUAAAGAUAAAGCUGGAAAAGGCAGAA---------AGAGCAAGAAGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGC ....((((((....(((((((.(...........((((......((....---------...))......)))).).)))))))..))))))..(((((((((......))))))))).. ( -30.90) >DroYak_CAF1 27065 109 - 1 -----------AGAACGGAGAUGAUAAAGAUAAAGCUGGAAAAGGCAGAAAGAGCAGAAAGAGCAAGAAGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGC -----------.(.(((((((.............(((......))).............((.((........)).)))))))))...)......(((((((((......))))))))).. ( -24.10) >consensus CAACGUCUGCAAGAAUGGAGAUGAUAAAGAUAAAGCUGGAAAAGGGAGAA_________AGAGCAAG_AGCAGCGCUUCUCCGUGGGCGGAUUGAGCGCAUAAUGAUGUUUAUGUGUUGC ...(((((.((....)).)))))...........(((........................))).....(((((((((.((((....))))..))))((((((......))))))))))) (-21.44 = -23.12 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:04 2006