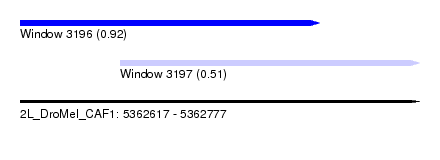
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,362,617 – 5,362,777 |
| Length | 160 |
| Max. P | 0.922197 |

| Location | 5,362,617 – 5,362,737 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -29.33 |
| Energy contribution | -29.25 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
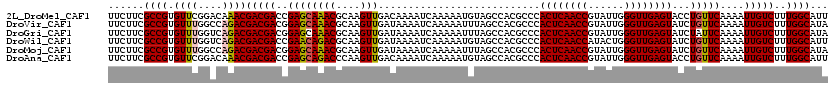
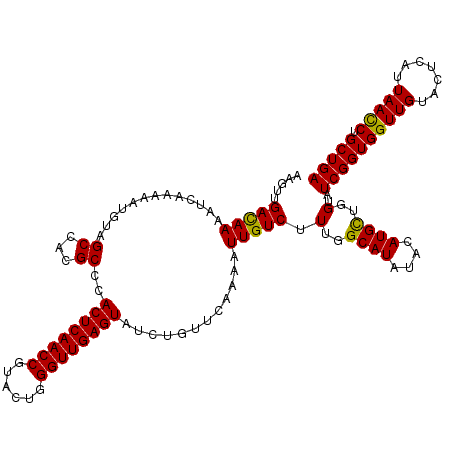
>2L_DroMel_CAF1 5362617 120 + 22407834 UUCUUCGCCGUGUUCGGACAAACGACGACCGAGCAAACGCAAGUUGACAAAAUCAAAAAUGUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUACCUGUUCAAAAUUGUCUUUGGCAUU ......(((((((((((.(.......).)))))))(((....)))(((((.............((...))..((((((((......))))))))............)))))..))))... ( -32.60) >DroVir_CAF1 43783 120 + 1 UUCUUCGCCGUGUUUGGCCAGACGACGACGGAGCAAACGCAAGUUGAUAAAAUCAAAAAUUUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUA ................((((((.(((((..((((((((....)))...........................((((((((......))))))))...)))))....)))))))))))... ( -33.90) >DroGri_CAF1 38376 120 + 1 UUCUUCGCCGUGUUUGGUCAGACGACGACGGAGCAAACGCAAGUUGAUAAAAUCAAAAAUUUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUAUCUAUUCAAAAUUGUCUUUGGCAUA ......((((.((((....))))(((((.((.((.(((....)))(((...)))..............))))((((((((......))))))))............)))))..))))... ( -31.40) >DroWil_CAF1 43499 120 + 1 UUCUUCGCCGUGUUUGGUCAGACGACGACCGAACAGACGCAAGUUGAUAAAAUCAAAAAUGUAGCCACGCCCACUCAACCAUACUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUU ......((((.....((((.......))))(((((((.(((..(((((...)))))...)))..........((((((((......)))))))).)))))))...........))))... ( -35.30) >DroMoj_CAF1 39353 120 + 1 UUCUUCGCCGUGUUUGGCCAGACGACGACGGAGCAAACGCAAGUUGAUAAAAUCAAAAAUUUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUA ................((((((.(((((..((((((((....)))...........................((((((((......))))))))...)))))....)))))))))))... ( -33.90) >DroAna_CAF1 26550 120 + 1 UUCUUCGCCGUGUUCGGACAAACGACGACCGAGCAGACCCAAGUUGACAAAAUCAAAAAUGUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUACCUGUUCAAAAUUGUCUUUGGCAUU ......(((((((((((.(.......).)))))))(((.....((((((..............((...))..((((((((......))))))))...)).))))....)))..))))... ( -30.20) >consensus UUCUUCGCCGUGUUUGGACAGACGACGACCGAGCAAACGCAAGUUGAUAAAAUCAAAAAUGUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUA ......((((.((((....))))(((((..((((((((....)))...........................((((((((......))))))))...)))))....)))))..))))... (-29.33 = -29.25 + -0.08)
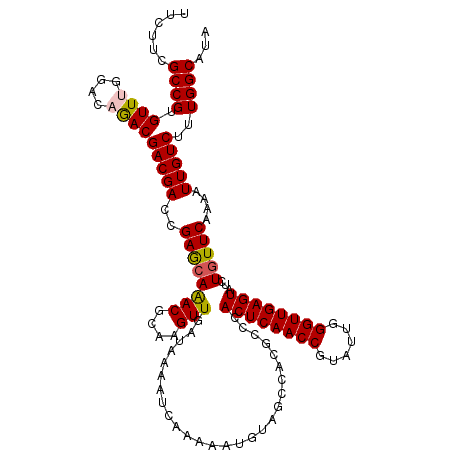
| Location | 5,362,657 – 5,362,777 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -27.25 |
| Energy contribution | -26.62 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5362657 120 + 22407834 AAGUUGACAAAAUCAAAAAUGUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUACCUGUUCAAAAUUGUCUUUGGCAUUUACAUGUUGGUGUCGGUGGUUGUGCUCAUUAACCUGCUGA ...((((.....)))).((((.(((.(((.((((((((((......))))))))...........((((.(.(..((((....))))..).).)))))).)))))))))).......... ( -28.70) >DroVir_CAF1 43823 120 + 1 AAGUUGAUAAAAUCAAAAAUUUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUAUACAUGCUGGUAUCGGUGGUUGUACUCAUUAACCUGCUGA ...(((((...))))).......((((.((....((((((......))))))((((.(((.((((......))))))).))))..)))))).((((((((((.......))))).))))) ( -29.10) >DroPse_CAF1 35968 120 + 1 AAGUUGACAAAAUCAAAAAUGUAGCCACGCCCACUCAACCGUACUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUAUACAUGCUGGUGUCGGUGGUUGUCCUAAUUAACCUGCUGA ...((((.....))))....(((((((((((.((((((((......))))))))....................(((((....)))))))))).)))(((((.......))))))))... ( -30.60) >DroGri_CAF1 38416 120 + 1 AAGUUGAUAAAAUCAAAAAUUUAGCCACGCCCACUCAACCGUAUUGGGUUGAGUAUCUAUUCAAAAUUGUCUUUGGCAUAUACAUGUUGGUAUCGGUGGUUGUACUCAUUAACCUGCUGA ...(((((...)))))....(((((...(((.((((((((......))))))))..............(...(..((((....))))..)...))))(((((.......))))).))))) ( -26.70) >DroWil_CAF1 43539 120 + 1 AAGUUGAUAAAAUCAAAAAUGUAGCCACGCCCACUCAACCAUACUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUUUACAUGUUGGUAUCGGUGGUUGUGCUCAUUAAUCUGCUGA ...(((((...))))).((((.(((.(((.((((((((((......))))))))...........((((...(..((((....))))..)...)))))).)))))))))).......... ( -28.00) >DroPer_CAF1 35752 120 + 1 AAGUUGACAAAAUCAAAAAUGUAGCCACGCCCACUCAACCGUACUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUAUACAUGCUGGUGUCGGUGGUUGUCCUAAUUAACCUGCUGA ...((((.....))))....(((((((((((.((((((((......))))))))....................(((((....)))))))))).)))(((((.......))))))))... ( -30.60) >consensus AAGUUGACAAAAUCAAAAAUGUAGCCACGCCCACUCAACCGUACUGGGUUGAGUAUCUGUUCAAAAUUGUCUUUGGCAUAUACAUGCUGGUAUCGGUGGUUGUACUCAUUAACCUGCUGA .....(((((.............((...))..((((((((......))))))))............))))).(..((((....))))..)..((((((((((.......))))).))))) (-27.25 = -26.62 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:25 2006