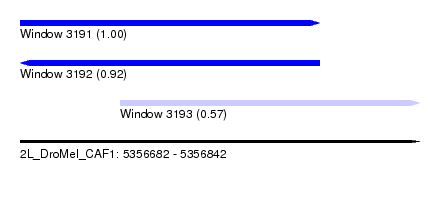
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,356,682 – 5,356,842 |
| Length | 160 |
| Max. P | 0.998714 |

| Location | 5,356,682 – 5,356,802 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -47.35 |
| Consensus MFE | -42.60 |
| Energy contribution | -42.79 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5356682 120 + 22407834 UGGCCGACAAGUUCAAGGCGAGCAUCUUCGAGCGCACCAAGGAUGGCAGCACGCUGAUGCACAUUGCGUCACUCAACGGUCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCU ((((......((((((((......)))).))))((((..((.(((((.......((((((.....)))))).......))))).))..)))))))).(((((((((....))))))))). ( -46.04) >DroVir_CAF1 36646 120 + 1 UGGCGGACAAAUUCAAGGCGAGCAUCUUCGAGCGCACCAAGGAUGGCAGCACACUGAUGCACAUUGCAUCACUUAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGUGUCU ....(((((..(((..((.(((((((...(.((((((..((.(((((.......((((((.....)))))).......))))).))..)))))))..))))))).))..)))...))))) ( -50.64) >DroGri_CAF1 30626 120 + 1 UGGCGGACAAGUUCAAAGCGAGCAUCUUCGAGCGCACCAAGGAUGGCAGCACUUUGAUGCACAUUGCAUCCCUGAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCU ((((......((((.(((.......))).))))((((..((.(((((........(((((.....)))))........))))).))..)))))))).(((((((((....))))))))). ( -47.49) >DroWil_CAF1 34047 120 + 1 UGGCGGACAAAUUCAAGGCGAGCAUCUUUGAGCGCACCAAGGAUGGCAGCACACUGAUGCACAUUGCAUCACUCAAUGGUCAUGCUGAGUGCGCCACUAUGCUCUUCAAGAAGGGUGUCU ....(((((..(((..((.((((((...((.((((((..((.(((((.......((((((.....)))))).......))))).))..))))))))..)))))).))..)))...))))) ( -46.24) >DroMoj_CAF1 32444 120 + 1 UGGCGGACAAAUUCAAGGCGAGCAUCUUCGAGCGCACCAAGGAUGGCAGCACACUGAUGCACAUCGCAUCACUCAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCU .((((..(...(((..((.(((((((...(.((((((..((.(((((.......((((((.....)))))).......))))).))..)))))))..))))))).))..))).).)))). ( -48.84) >DroAna_CAF1 20379 120 + 1 UGGCCGACAAGUUCAAGGCGAGCAUCUUCGAGCGCACCAAGGAUGGCAGCACUCUGAUGCACAUUGCGUCACUCAACGGUCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUGU ..(((((.....))..((.(((((((...(.((((((..((.(((((.......((((((.....)))))).......))))).))..)))))))..))))))).))......))).... ( -44.84) >consensus UGGCGGACAAAUUCAAGGCGAGCAUCUUCGAGCGCACCAAGGAUGGCAGCACACUGAUGCACAUUGCAUCACUCAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCU .....(((...(((..((.(((((((...(.((((((..((.(((((.......((((((.....)))))).......))))).))..)))))))..))))))).))..)))....))). (-42.60 = -42.79 + 0.19)

| Location | 5,356,682 – 5,356,802 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -47.07 |
| Consensus MFE | -39.04 |
| Energy contribution | -39.35 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5356682 120 - 22407834 AGACGCCCUUCUUGAAGAGCAUCGUGGCGCACUCAGCAUGACCGUUGAGUGACGCAAUGUGCAUCAGCGUGCUGCCAUCCUUGGUGCGCUCGAAGAUGCUCGCCUUGAACUUGUCGGCCA .((((...(((..(..(((((((...(((((((((((......)))))))).)))...(((((((((.(((....))).).)))))))).....)))))))..)..)))..))))..... ( -46.60) >DroVir_CAF1 36646 120 - 1 AGACACCCUUCUUGAAGAGCAUCGUGGCGCACUCAGCAUGGCCAUUAAGUGAUGCAAUGUGCAUCAGUGUGCUGCCAUCCUUGGUGCGCUCGAAGAUGCUCGCCUUGAAUUUGUCCGCCA .((((...(((..(..(((((((..((((((((.((.(((((.....(.((((((.....)))))).).....))))).)).))))))))....)))))))..)..)))..))))..... ( -50.40) >DroGri_CAF1 30626 120 - 1 AGACGCCCUUCUUGAAGAGCAUCGUGGCGCACUCAGCAUGGCCAUUCAGGGAUGCAAUGUGCAUCAAAGUGCUGCCAUCCUUGGUGCGCUCGAAGAUGCUCGCUUUGAACUUGUCCGCCA .((((...(((.....(((((((..((((((((.((.(((((((((....(((((.....)))))..))))..))))).)).))))))))....))))))).....)))..))))..... ( -47.60) >DroWil_CAF1 34047 120 - 1 AGACACCCUUCUUGAAGAGCAUAGUGGCGCACUCAGCAUGACCAUUGAGUGAUGCAAUGUGCAUCAGUGUGCUGCCAUCCUUGGUGCGCUCAAAGAUGCUCGCCUUGAAUUUGUCCGCCA .((((...(((..(..((((((..(((((((((((((((........(.((((((.....)))))).)))))))........))))))).))...))))))..)..)))..))))..... ( -41.70) >DroMoj_CAF1 32444 120 - 1 AGACGCCCUUCUUGAAGAGCAUCGUGGCGCACUCAGCAUGGCCAUUGAGUGAUGCGAUGUGCAUCAGUGUGCUGCCAUCCUUGGUGCGCUCGAAGAUGCUCGCCUUGAAUUUGUCCGCCA .((((...(((..(..(((((((..((((((((.((.((((((((..(.((((((.....)))))).))))..))))).)).))))))))....)))))))..)..)))..))))..... ( -48.00) >DroAna_CAF1 20379 120 - 1 ACACGCCCUUCUUGAAGAGCAUCGUGGCGCACUCAGCAUGACCGUUGAGUGACGCAAUGUGCAUCAGAGUGCUGCCAUCCUUGGUGCGCUCGAAGAUGCUCGCCUUGAACUUGUCGGCCA ....(((.(((..(..(((((((...(((((((((((......)))))))).)))...........((((((.((((....))))))))))...)))))))..)..)))......))).. ( -48.10) >consensus AGACGCCCUUCUUGAAGAGCAUCGUGGCGCACUCAGCAUGACCAUUGAGUGAUGCAAUGUGCAUCAGUGUGCUGCCAUCCUUGGUGCGCUCGAAGAUGCUCGCCUUGAACUUGUCCGCCA .((((...(((..(..(((((((..((((((((((((((.((......))(((((.....)))))...))))))........))))))))....)))))))..)..)))..))))..... (-39.04 = -39.35 + 0.31)
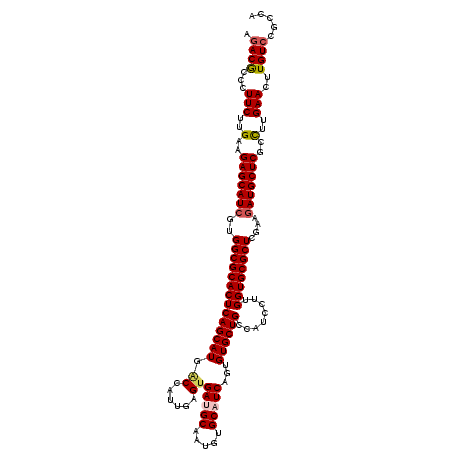
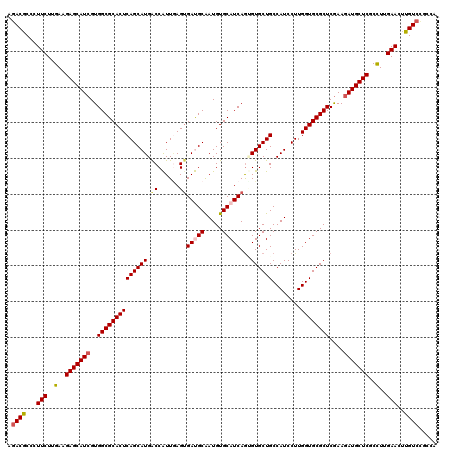
| Location | 5,356,722 – 5,356,842 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -34.92 |
| Energy contribution | -34.92 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5356722 120 + 22407834 GGAUGGCAGCACGCUGAUGCACAUUGCGUCACUCAACGGUCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCUACCUCCAUAUGCCCAACAAGGAUGGAGCCCGGAGUAUUCA (.(((((.......((((((.....)))))).......))))).).((((((.((..(((((((((....)))))))))...((((((...((......))))))))...)).)))))). ( -43.44) >DroVir_CAF1 36686 120 + 1 GGAUGGCAGCACACUGAUGCACAUUGCAUCACUUAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGUGUCUAUCUUCACAUGCCCAACAAGGAUGGCGCCCGCAGCAUACA ..(((((.......((((((.....)))))).......)))))((((.(.((((((.(((((((((....))))))))).............((.....)).)))))).).))))..... ( -44.84) >DroGri_CAF1 30666 120 + 1 GGAUGGCAGCACUUUGAUGCACAUUGCAUCCCUGAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCUACCUCCACAUGCCCAACAAGGAUGGGGCACGCAGCAUACA ..(((((........(((((.....)))))........)))))((((.((((.(((.(((((((((....)))))))))....(((.............)))))).)))).))))..... ( -46.61) >DroWil_CAF1 34087 120 + 1 GGAUGGCAGCACACUGAUGCACAUUGCAUCACUCAAUGGUCAUGCUGAGUGCGCCACUAUGCUCUUCAAGAAGGGUGUCUAUCUGCAUAUGCCCAAUAAGGAUGGAGCACGUAGCAUACA ..(((((.......((((((.....)))))).......)))))((((.((((........((((((....)))))).(((((((...(((.....))).))))))))))).))))..... ( -36.64) >DroMoj_CAF1 32484 120 + 1 GGAUGGCAGCACACUGAUGCACAUCGCAUCACUCAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCUAUCUGCACAUGCCCAACAAGGAUGGCGCCCGCAGCAUACA ..(((((.......((((((.....)))))).......)))))((((.(.((((((.(((((((((....))))))))).............((.....)).)))))).).))))..... ( -47.14) >DroAna_CAF1 20419 120 + 1 GGAUGGCAGCACUCUGAUGCACAUUGCGUCACUCAACGGUCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUGUACCUCCACAUGCCCAACAAGGACGGAGCCCGAAGUAUCCA (((((((.((((((((((((.....))))))......((.....))))))))))).((..(((((((..(..((((((((......))))))))..)...)).))))).))....)))). ( -47.60) >consensus GGAUGGCAGCACACUGAUGCACAUUGCAUCACUCAAUGGCCAUGCUGAGUGCGCCACGAUGCUCUUCAAGAAGGGCGUCUACCUCCACAUGCCCAACAAGGAUGGAGCCCGCAGCAUACA ..(((((.......((((((.....)))))).......)))))((((.(.((.(((.(((((((((....))))))))).............((.....)).))).)).).))))..... (-34.92 = -34.92 + 0.00)


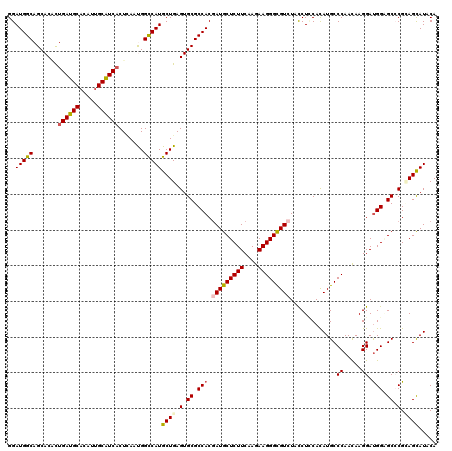
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:21 2006