
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,356,150 – 5,356,264 |
| Length | 114 |
| Max. P | 0.725862 |

| Location | 5,356,150 – 5,356,264 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -40.05 |
| Consensus MFE | -32.72 |
| Energy contribution | -32.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
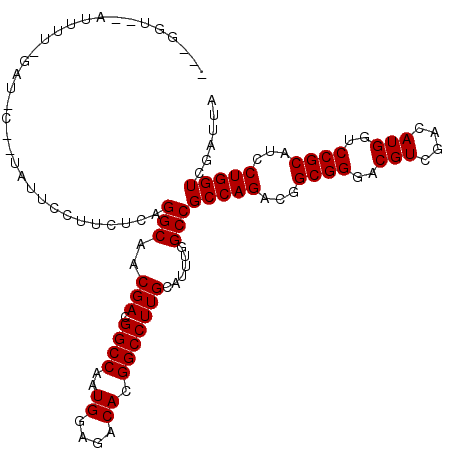
>2L_DroMel_CAF1 5356150 114 + 22407834 CAUGUUAAAUUUUGGAU-U--UAUUUUUUCGCAGGCAACGACGGCCAAUGGAGACACGGCCUUGCAUUUGGCCGCCAGACGGCGGGACGUGGACAUGGUCCGCAUCCUGGUUGAUUA .................-.--.........((((....)...((((..((....)).)))).)))..(..((((((....)))((((.((((((...)))))).)))))))..)... ( -38.90) >DroVir_CAF1 36279 114 + 1 ---CGUGCAUUCGUGAUUCGUUAUUCGUGAUAAGGCAACGACGGCCAAUGGAGACACGGCCUUGCAUUUGGCCGCCAGACGGCGGGACGUCGAUAUGGUCCGCAUUCUGGUUGAUUA ---.(((..(((((...((((..((.((......))))..))))...)))))..)))((((........))))((((((..((((..(((....)))..))))..))))))...... ( -38.80) >DroGri_CAF1 30274 101 + 1 ---GGU------------CG-AUUUCCCUUUCAGGCAACGACGGCCAAUGGAGACACGGCCUUGCAUUUGGCCGCCAGACGGCGGGACGUCGAUAUGGUCCGCAUCCUGGUCGACUA ---.((------------((-((.((((.....(((..(((.((((..((....)).)))))))......)))(((....))))))).)))))).(((((.((......)).))))) ( -39.50) >DroSim_CAF1 14477 106 + 1 --------AUUUUGGAU-U--UAUUUUUUCGCAGGCAACGACGGCCAAUGGAGACACGGCCUUGCAUUUGGCCGCCAGACGGCGGGACGUGGACAUGGUCCGCAUCCUGGUCGAUUA --------.........-.--.........((((....)...((((..((....)).)))).)))..(((((((((....)))((((.((((((...)))))).))))))))))... ( -40.20) >DroMoj_CAF1 32080 100 + 1 ---UUU--------------CUUUUCGAUUUUAGGCAACGACGGCCAAUGGAGACACGGCCUUGCAUUUGGCCGCCAGACGGCGGGACGUCGACAUGGUCCGCAUUCUGGUGGAUUA ---...--------------..............((((....((((..((....)).))))))))......((((((((..((((..(((....)))..))))..)))))))).... ( -37.40) >DroAna_CAF1 19665 114 + 1 CAUGCUCGUAUUUCGUU-C--CCUUUCCCCCCAGGCAACGACGGCCAAUGGAGACACGGCCUUGCAUUUGGCCGCCAGACGGCGGGACGUGGACAUGGUCCGCAUCCUGGUCGACUA .((((.......(((((-.--(((........))).))))).((((..((....)).))))..))))(((((((((....)))((((.((((((...)))))).))))))))))... ( -45.50) >consensus ___GGU__AUUUU_GAU_C__UAUUCCUUCUCAGGCAACGACGGCCAAUGGAGACACGGCCUUGCAUUUGGCCGCCAGACGGCGGGACGUCGACAUGGUCCGCAUCCUGGUCGAUUA .................................(((..(((.((((..((....)).)))))))......)))(((((...((((..(((....)))..))))...)))))...... (-32.72 = -32.72 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:17 2006