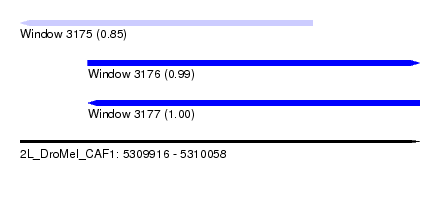
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,309,916 – 5,310,058 |
| Length | 142 |
| Max. P | 0.998994 |

| Location | 5,309,916 – 5,310,020 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -30.40 |
| Energy contribution | -31.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
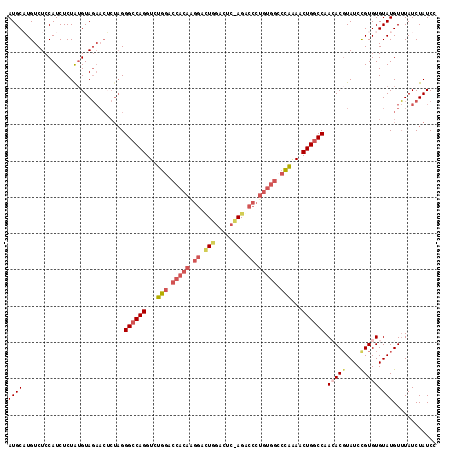
>2L_DroMel_CAF1 5309916 104 - 22407834 UUCGGGCCACAGGGUCUUGUGUCAAAUCCUUGUGGUCCAGACCUGGCCCUAGAGUUCUACAUAGAGAUGGAGACAUGCAUGCAUCGGUAUCUAAUAGUUAUCAC ...(((((((((((..(((...)))..)))))))))))..(((..((.......((((....))))(((....)))....))...)))................ ( -29.90) >DroSec_CAF1 85620 104 - 1 UUUGGUCCACAGGGCCUGGAGUCCAGUCCCUGUGGUCCAGACCUGGCCCUAGAGUUCUACAUAGAGAUGGAGACAUGCAUGCAUCGGUAUCUUAUAGUAAACAC (((((.((((((((.((((...)))).)))))))).)))))............(((.(((...((((((..((..((....))))..))))))...)))))).. ( -35.80) >DroSim_CAF1 89427 103 - 1 UUUGGGCCACAGGGCCUGGAGUCCAGUCCCUGUGGUCCAGACCUGGCCCUAGAGUUCUAC-UAGAGAUGGAGACAUGCAUGCAUUGGUAUCUUAUAGUAAACAC .(((((((((((((.((((...)))).)))))))))))))(((..((.((((.......)-)))..(((....)))....))...)))................ ( -41.20) >DroYak_CAF1 90376 102 - 1 UUUAGGCCACAGGGUCU--AUCCGAGUCCUUGUGGUCCAGACCUGGCCCUAGCGUUCUACACAGAGAUGGAGACAAGCAUGCAUCGGUAUCUAAUAGUAAACAC ....((((((((((.((--.....)).)))))))))).(((....(((...(((((((......)))((....))...))))...))).)))............ ( -27.70) >consensus UUUGGGCCACAGGGCCUGGAGUCCAGUCCCUGUGGUCCAGACCUGGCCCUAGAGUUCUACAUAGAGAUGGAGACAUGCAUGCAUCGGUAUCUAAUAGUAAACAC .(((((((((((((.(((.....))).)))))))))))))(((..((.......((((....))))(((....)))....))...)))................ (-30.40 = -31.40 + 1.00)

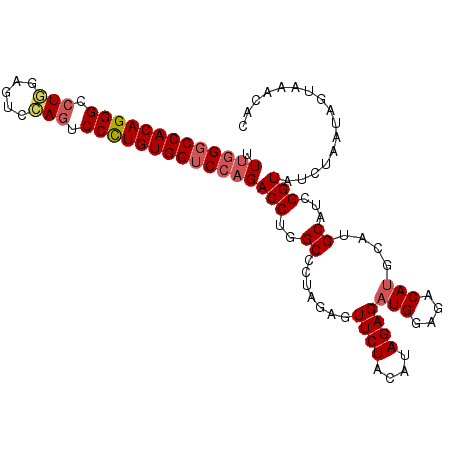
| Location | 5,309,940 – 5,310,058 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -23.86 |
| Energy contribution | -25.96 |
| Covariance contribution | 2.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
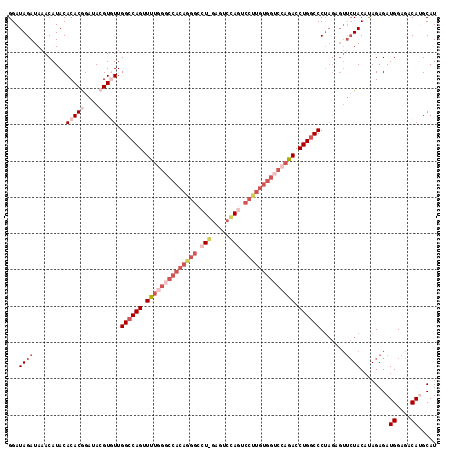
>2L_DroMel_CAF1 5309940 118 + 22407834 AUGCAUGUCUCCAUCUCUAUGUAGAACUCUAGGGCCAGGUCUGGACCACAAGGAUUUGACACAAGACCCUGUGGCCCGAAACUGGCCAACACUUAUCUGUGUGUAUGUUUAUCUAUCC .((((((..........))))))((((.....((((((...(((.(((((.((.((((...)))).)).))))).)))...)))))).((((......))))....))))........ ( -34.50) >DroSec_CAF1 85644 118 + 1 AUGCAUGUCUCCAUCUCUAUGUAGAACUCUAGGGCCAGGUCUGGACCACAGGGACUGGACUCCAGGCCCUGUGGACCAAAACUGUCCAACACGUAUCCGUGUGUAUGUUUAUCUAUCC .((((((..........))))))((((....((((.((...(((.((((((((.((((...)))).)))))))).)))...)))))).(((((....)))))....))))........ ( -40.80) >DroSim_CAF1 89451 117 + 1 AUGCAUGUCUCCAUCUCUA-GUAGAACUCUAGGGCCAGGUCUGGACCACAGGGACUGGACUCCAGGCCCUGUGGCCCAAAACUGGCCAACACGUAUCCGUGUGUAUGUUUAUCUAUCC ..(((((.........(((-(.......))))((((((...(((.((((((((.((((...)))).)))))))).)))...)))))).(((((....))))).))))).......... ( -48.20) >DroEre_CAF1 83126 90 + 1 AUGCUUGUCUCCAUCUCUAUGUAU-------GGGCCAGGUCUGGACCAUAAGGAC---------------------UAGACCUGGCCAAAACGUAUCUGUGUGUAUGUGUAUCUAUCC ((((.......(((...(((((..-------.((((((((((((.((....)).)---------------------)))))))))))...)))))...))).......))))...... ( -32.54) >DroYak_CAF1 90400 116 + 1 AUGCUUGUCUCCAUCUCUGUGUAGAACGCUAGGGCCAGGUCUGGACCACAAGGACUCGGAU--AGACCCUGUGGCCUAAACCUGGCCAAGACGUAUCCGUGUGUAUGUUUAUCUAUCC ..((...(((.(((....))).)))..))(((((((((((.(((.(((((.((.((.....--)).)).))))).))).)))))))).((((((((......))))))))..)))... ( -39.10) >consensus AUGCAUGUCUCCAUCUCUAUGUAGAACUCUAGGGCCAGGUCUGGACCACAAGGACUGGACUC_AGACCCUGUGGCCCAAAACUGGCCAACACGUAUCCGUGUGUAUGUUUAUCUAUCC ((((............................((((((...(((.(((((.((.(((.....))).)).))))).)))...)))))).(((((....)))))))))............ (-23.86 = -25.96 + 2.10)

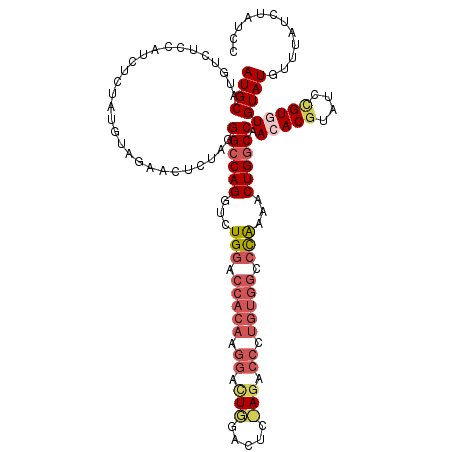
| Location | 5,309,940 – 5,310,058 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -31.88 |
| Energy contribution | -36.30 |
| Covariance contribution | 4.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
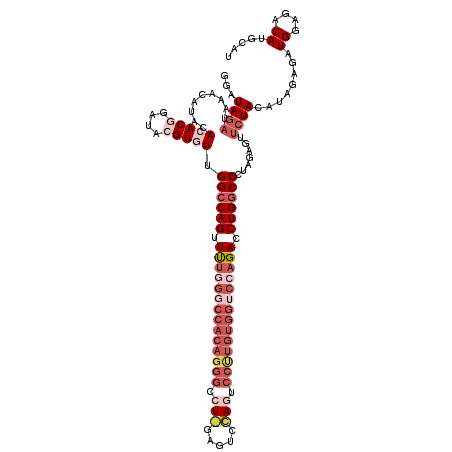
>2L_DroMel_CAF1 5309940 118 - 22407834 GGAUAGAUAAACAUACACACAGAUAAGUGUUGGCCAGUUUCGGGCCACAGGGUCUUGUGUCAAAUCCUUGUGGUCCAGACCUGGCCCUAGAGUUCUACAUAGAGAUGGAGACAUGCAU ...((((.........((((......)))).((((((.(..(((((((((((..(((...)))..)))))))))))..).)))))).......)))).......(((....))).... ( -41.50) >DroSec_CAF1 85644 118 - 1 GGAUAGAUAAACAUACACACGGAUACGUGUUGGACAGUUUUGGUCCACAGGGCCUGGAGUCCAGUCCCUGUGGUCCAGACCUGGCCCUAGAGUUCUACAUAGAGAUGGAGACAUGCAU ...((((.........(((((....))))).((.(((.(((((.((((((((.((((...)))).)))))))).))))).)))..))......)))).......(((....))).... ( -45.80) >DroSim_CAF1 89451 117 - 1 GGAUAGAUAAACAUACACACGGAUACGUGUUGGCCAGUUUUGGGCCACAGGGCCUGGAGUCCAGUCCCUGUGGUCCAGACCUGGCCCUAGAGUUCUAC-UAGAGAUGGAGACAUGCAU ((.((((.........(((((....))))).((((((.((((((((((((((.((((...)))).)))))))))))))).)))))).......)))))-)....(((....))).... ( -56.40) >DroEre_CAF1 83126 90 - 1 GGAUAGAUACACAUACACACAGAUACGUUUUGGCCAGGUCUA---------------------GUCCUUAUGGUCCAGACCUGGCCC-------AUACAUAGAGAUGGAGACAAGCAU ..........................((...((((((((((.---------------------(.((....)).).)))))))))).-------..)).......((....))..... ( -28.20) >DroYak_CAF1 90400 116 - 1 GGAUAGAUAAACAUACACACGGAUACGUCUUGGCCAGGUUUAGGCCACAGGGUCU--AUCCGAGUCCUUGUGGUCCAGACCUGGCCCUAGCGUUCUACACAGAGAUGGAGACAAGCAU ....................(((.((((...((((((((((.((((((((((.((--.....)).)))))))))).))))))))))...))))))).........((....))..... ( -47.00) >consensus GGAUAGAUAAACAUACACACGGAUACGUGUUGGCCAGUUUUGGGCCACAGGGCCU_GAGUCCAGUCCUUGUGGUCCAGACCUGGCCCUAGAGUUCUACAUAGAGAUGGAGACAUGCAU ...((((.........(((((....))))).((((((.((((((((((((((.(((.....))).)))))))))))))).)))))).......))))........((....))..... (-31.88 = -36.30 + 4.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:05 2006