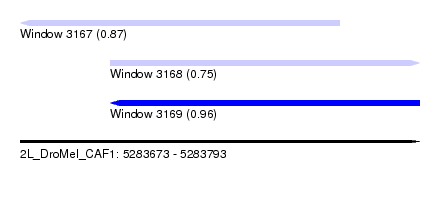
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,283,673 – 5,283,793 |
| Length | 120 |
| Max. P | 0.960076 |

| Location | 5,283,673 – 5,283,769 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -14.93 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
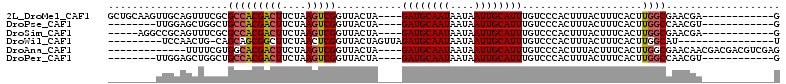
>2L_DroMel_CAF1 5283673 96 - 22407834 GCUGCAAGUUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUA----GAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAACGA------------G (((((.....))))).(((((((((((((....)))))(..((.(----((((((((....)))))))))))..)..............)))))..)))------------. ( -29.90) >DroPse_CAF1 67639 88 - 1 --------UUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUA----GAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCCAACGU------------G --------.....(.(((((....(((((....)))))(..((.(----((((((((....)))))))))))..)...............))))).)..------------. ( -22.20) >DroSim_CAF1 65590 91 - 1 -----AGGCCGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUA----GAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAACGA------------G -----...........(((((((((((((....)))))(..((.(----((((((((....)))))))))))..)..............)))))..)))------------. ( -24.00) >DroWil_CAF1 15190 86 - 1 ---------UCCAACUG-CAGCAGCGGCUUCUAACUCGGUUACUAGUUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCAU----------------U ---------.(((((((-(....))))..(((((((.(....).)))))))((((((....)))))).....................))))...----------------. ( -17.50) >DroAna_CAF1 44533 95 - 1 -------------UUUUCGUGGCACGACUUCUAAGUCGGUUACUA----GAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAACAACGACGACGUCGAG -------------.....(((((.(((((....))))))))))((----((((((((....))))))))))................(((((((...........))))))) ( -25.90) >DroPer_CAF1 69203 88 - 1 --------UUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUA----GAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCCAACGU------------G --------.....(.(((((....(((((....)))))(..((.(----((((((((....)))))))))))..)...............))))).)..------------. ( -22.20) >consensus ________UUGCAGCUGCCUGCCACGACUUCUAAGUCGGUUACUA____GAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAACGA____________G ....................(((((((((....)))))...........((((((((....))))))))....................))))................... (-14.93 = -15.15 + 0.22)
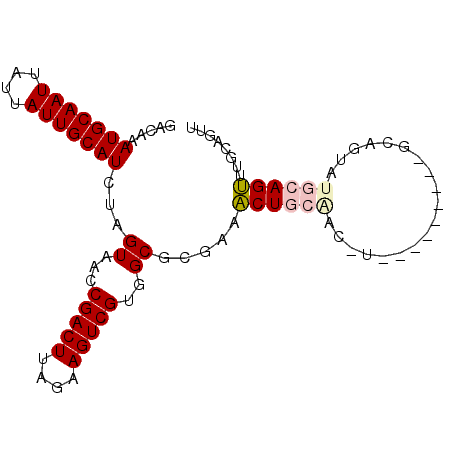
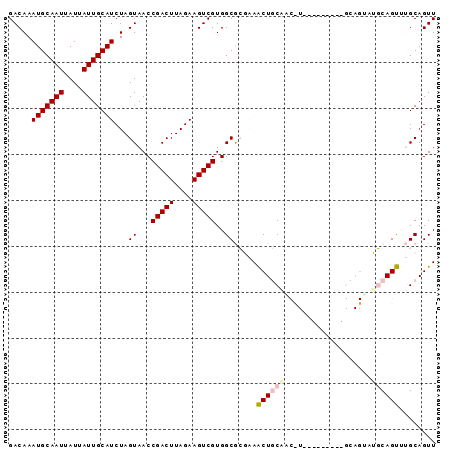
| Location | 5,283,700 – 5,283,793 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5283700 93 + 22407834 GACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAACUUGCAGCCUAUGCAGUAUGCAGUAUGCAGUU .....(((((((....)))))))...((...(((((....)))))..))(((..(((((((((.(((.....)))))).)))))).))).... ( -27.60) >DroPse_CAF1 67666 76 + 1 GACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCAGCCAGCUCCAA-----------------AGCCAGCCAGCAGUU (((..(((((((....)))))))...((...(((((....)))))((((.....(((....-----------------)))..)))))).))) ( -16.80) >DroSec_CAF1 59327 84 + 1 GACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAGCCU---------GCAGUAUGCAGUUUGCAGUU .....(((((((....)))))))...((..((((((....))))).)..))..((((((((.((---------((.....)))).)))))))) ( -29.30) >DroSim_CAF1 65617 84 + 1 GACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCGGCCU---------GCAGUAUGCAGUUUGCAGUU .....(((((((....)))))))...((..((((((....))))).)..))..(((((((..((---------((.....))))..))))))) ( -28.60) >DroEre_CAF1 55283 91 + 1 GACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAGCUUGCGGCCU--GCAGCUUGCAGUUUGCAGUU .....(((((((....)))))))...((...(((((....)))))..))((.(((((((((((.((.....--))))).)))))))))).... ( -30.50) >DroPer_CAF1 69230 76 + 1 GACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCAGCCAGCUCCAA-----------------AGCCAGCCAGCAGUU (((..(((((((....)))))))...((...(((((....)))))((((.....(((....-----------------)))..)))))).))) ( -16.80) >consensus GACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAAC_U_________GCAGUAUGCAGUUUGCAGUU .....(((((((....)))))))...((...(((((....)))))..)).....((((((...................))))))........ (-14.80 = -15.28 + 0.47)

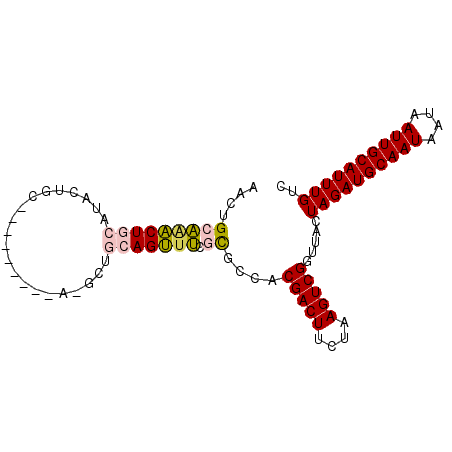
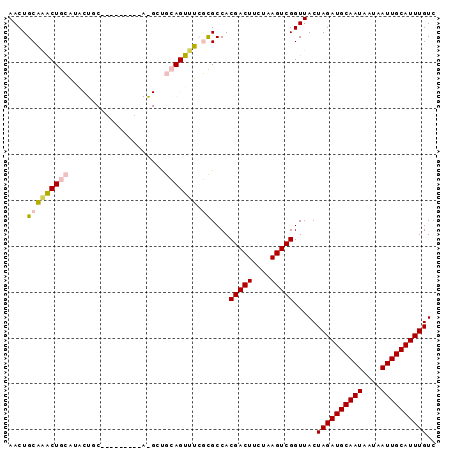
| Location | 5,283,700 – 5,283,793 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.15 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
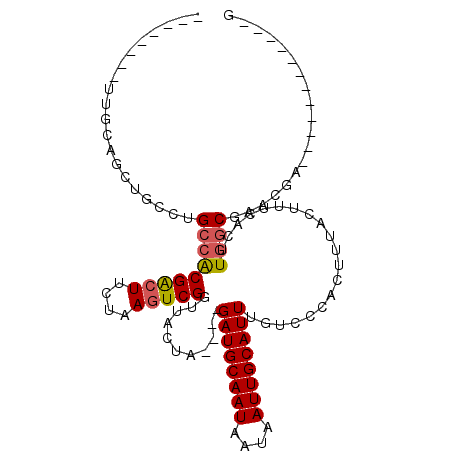
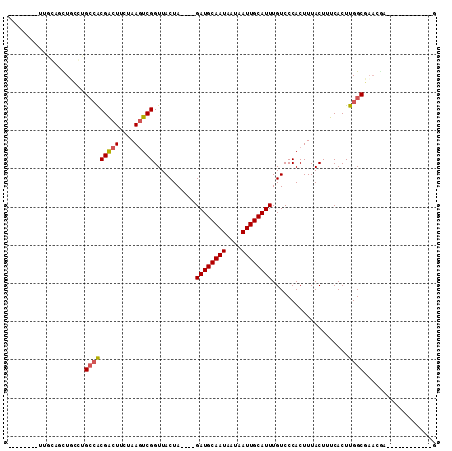
>2L_DroMel_CAF1 5283700 93 - 22407834 AACUGCAUACUGCAUACUGCAUAGGCUGCAAGUUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUC ....((..((((((.((((((.....))).)))))))))...))....(((((....))))).....((((((((((....)))))))))).. ( -28.90) >DroPse_CAF1 67666 76 - 1 AACUGCUGGCUGGCU-----------------UUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUC ....((.(((..(((-----------------....)))..)))))..(((((....))))).....((((((((((....)))))))))).. ( -26.20) >DroSec_CAF1 59327 84 - 1 AACUGCAAACUGCAUACUGC---------AGGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUC (((((((..((((.....))---------))..)))))))........(((((....))))).....((((((((((....)))))))))).. ( -30.50) >DroSim_CAF1 65617 84 - 1 AACUGCAAACUGCAUACUGC---------AGGCCGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUC ((((((...((((.....))---------))...))))))........(((((....))))).....((((((((((....)))))))))).. ( -27.50) >DroEre_CAF1 55283 91 - 1 AACUGCAAACUGCAAGCUGC--AGGCCGCAAGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUC ....((((((((((.(((((--.....)).))))))))))).))....(((((....))))).....((((((((((....)))))))))).. ( -32.80) >DroPer_CAF1 69230 76 - 1 AACUGCUGGCUGGCU-----------------UUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUC ....((.(((..(((-----------------....)))..)))))..(((((....))))).....((((((((((....)))))))))).. ( -26.20) >consensus AACUGCAAACUGCAUACUGC_________A_GCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUC ....(((((((((.....................))))))).))....(((((....))))).....((((((((((....)))))))))).. (-19.26 = -19.15 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:58 2006