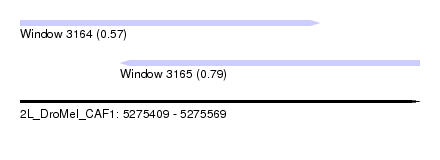
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,275,409 – 5,275,569 |
| Length | 160 |
| Max. P | 0.791868 |

| Location | 5,275,409 – 5,275,529 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -47.79 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.97 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
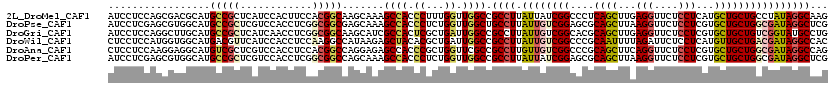
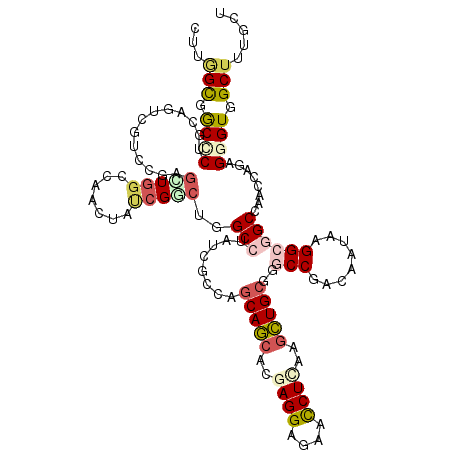
>2L_DroMel_CAF1 5275409 120 + 22407834 AUCCUCCAGCGACGCAUGCCGCUCAUCCACUUCCACGGCAAGCAAAGCCACCCUUUGGUUGGCCGCCUUAUUAUCGGCCCUCAGCUUGAGGUUCUCCUCAUGCUGCUGCCUAUAGGCAAG ........(((..((.(((((..............))))).))...((((((....)).)))))))(((.((((.(((...((((.(((((....))))).))))..))).))))..))) ( -39.84) >DroPse_CAF1 55365 120 + 1 AUCCUCGAGCGUGGCAUGCCGCUCGUCCACCUCGGCGGCGAGCAAAGCCACCCUCUGGUUGGCUGCCUUAUUGUCGGAGCGCAGCUUAAGGUUCUCCUCGUGCUGCUGGCGAUAGGCUCG .....(((((..((((.((((((((.((.(....).)))))))..(((((.....))))))))))))(((((((((....(((((...(((....)))...))))))))))))))))))) ( -53.80) >DroGri_CAF1 3333 120 + 1 AUCCUCCAGGCUUGCAUGCCGCUCAUCAACCUCGGCGGCAAGCAUCGCCACUCGCUGAUUGGCCGCCUUAUUGUCGGCACGCAGCUUGAGGUUCUCCUCGUGCUGCUGUCGGUAUGCCUG ......(((((.(((.(((((((..........))))))).))).........((((((..((((.(.....).))))..(((((.(((((....))))).))))).))))))..))))) ( -51.60) >DroWil_CAF1 6445 120 + 1 CUCCUCCAUGGUGGCAUGACGUUCAUCCACCUCCAAGGCCAUAAGAGCUACACGCUGAUUGGCCGCCUUAUUGUCGGCCCGCAAUUUUAGAUUCUCCUCAUGUUGCUGACGAUAGGCCAC .........(((((.(((.....)))))))).....(((((....(((.....)))...)))))((((.(((((((((..(((.................))).)))))))))))))... ( -39.83) >DroAna_CAF1 31182 120 + 1 CUCCUCCAAGGAGGCAUGUCGCUCGUCCACCUCCACGGCCAGGAGAGCCACCCGCUGGUUCGCCGCCUUGUUGUCGGCCCGCAGCUUCAGGUUCUCCUCGUGCUGCUGGCGAUAGGCCAG .........(((((.(((.....)))...)))))..((((.((.((((((.....)))))).)).......(((((.((.(((((...(((....)))...))))).))))))))))).. ( -48.90) >DroPer_CAF1 55376 120 + 1 AUCCUCGAGCGUGGCAUGCCGCUCGUCCACCUCGGCGGCCAGCAAAGCCACCCUCUGGUUGGCCGCCUUAUUAUCGGAGCGCAGCUUAAGGUUCUCCUCGUGCUGCUGGCGAUAGGCUCG .....(((((.((...((((((((.........((((((((((..((.....))...)))))))))).........))))(((((...(((....)))...))))).)))).)).))))) ( -52.77) >consensus AUCCUCCAGCGUGGCAUGCCGCUCAUCCACCUCCACGGCCAGCAAAGCCACCCGCUGGUUGGCCGCCUUAUUGUCGGCCCGCAGCUUAAGGUUCUCCUCGUGCUGCUGGCGAUAGGCCAG .................(((((............))))).......((((.((...)).)))).((((.((((((((....((((...(((....)))...))))))))))))))))... (-25.18 = -25.97 + 0.78)

| Location | 5,275,449 – 5,275,569 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -49.43 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.93 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
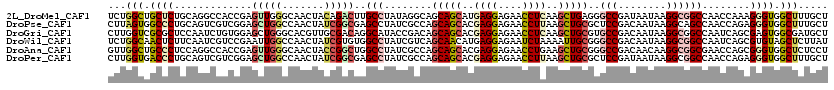
>2L_DroMel_CAF1 5275449 120 - 22407834 UCUGGCUGCUCUGCAGGCCACCGAGUUGGGCAACUACAGACUUGCCUAUAGGCAGCAGCAUGAGGAGAACCUCAAGCUGAGGGCCGAUAAUAAGGCGGCCAACCAAAGGGUGGCUUUGCU ...(((..((((...((((.((....(((((((........)))))))..(((..((((.(((((....))))).))))...)))........)).))))......))))..)))..... ( -51.00) >DroPse_CAF1 55405 120 - 1 CUUAGUGGCCCUGCAGUCGUCGGAGCUGGCCAACUAUCGGCGAGCCUAUCGCCAGCAGCACGAGGAGAACCUUAAGCUGCGCUCCGACAAUAAGGCAGCCAACCAGAGGGUGGCUUUGCU (..(((.(((((......((((((((............(((((.....))))).(((((..((((....))))..))))))))))))).....((.......))..))))).)))..).. ( -51.90) >DroGri_CAF1 3373 120 - 1 CUUGGUCGCGCUCCAAUCUGUGGAGCUGGGCACGUUGCGACAGGCAUACCGACAGCAGCACGAGGAGAACCUCAAGCUGCGUGCCGACAAUAAGGCGGCCAAUCAGCGAGUGGCGAUGCU ..(((((((((((((.....))))))..((((((((((.....))).)).....(((((..((((....))))..)))))))))).........)))))))(((.((.....)))))... ( -50.90) >DroWil_CAF1 6485 120 - 1 UCUGGCAACUCUUCAAUCGUCCGAAUUGGCCAACUAUCGUGUGGCCUAUCGUCAGCAACAUGAGGAGAAUCUAAAAUUGCGGGCCGACAAUAAGGCGGCCAAUCAGCGUGUAGCUCUUAU ...(((..(((((((...((.(((...(((((((....)).)))))..)))...))....)))))))...........((.(((((.(......)))))).....)).....)))..... ( -32.60) >DroAna_CAF1 31222 120 - 1 GUUGGCUGCCCUCCAGGCCACCGAGUUGGGCAACUACCGGCUGGCCUAUCGCCAGCAGCACGAGGAGAACCUGAAGCUGCGGGCCGACAACAAGGCGGCGAACCAGCGGGUGGCUCUCCU (..(((..(((...(((((((((.((.((....))))))).)))))).(((((.(((((...(((....)))...)))))..(((........))))))))......)))..)))..).. ( -58.10) >DroPer_CAF1 55416 120 - 1 CUUGGUGACCCUGCAGUCGUCGGAGCUGGCCAACUAUCGGCGAGCCUAUCGCCAGCAGCACGAGGAGAACCUUAAGCUGCGCUCCGAUAAUAAGGCGGCCAACCAGAGGGUGGCUUUGCU (..(((.(((((((.((..(((((((............(((((.....))))).(((((..((((....))))..))))))))))))..))...))((....))..))))).)))..).. ( -52.10) >consensus CUUGGCGGCCCUGCAGUCGUCCGAGCUGGCCAACUAUCGGCUGGCCUAUCGCCAGCAGCACGAGGAGAACCUCAAGCUGCGGGCCGACAAUAAGGCGGCCAACCAGAGGGUGGCUUUGCU ...(((.((((.............(((((.......)))))..(((........(((((..((((....))))..)))))..(((........))))))........)))).)))..... (-23.82 = -24.93 + 1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:54 2006