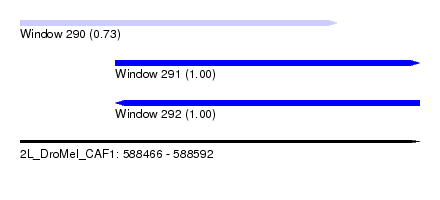
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 588,466 – 588,592 |
| Length | 126 |
| Max. P | 0.999999 |

| Location | 588,466 – 588,566 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -18.36 |
| Energy contribution | -17.43 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 588466 100 + 22407834 CGUAAAAU----------AUAUGAUUAAAAGGAAAAAAAGCAACCCGCUUGUUUGUAUGCAUGCAUGUGUGUGUGACGUGCCAAUUUGCAUGGCGUUUCGUGCAUACAAA .(((...(----------(((..(.(((..((............))..))).)..))))..))).((((((..(((..(((((.......)))))..)))..)))))).. ( -25.40) >DroSec_CAF1 19773 100 + 1 CGUACAAU----------AUAUGAUUAAAAGGGGAAAAAACAACCCGCCUAUUUGUAUGUAUGUAUGUGUGAGUGACGCGCCAACUUGCAUGGCAUUUCGUACAUACAAA ........----------............(((..........))).....((((((((((((...((((.....))))((((.......))))....)))))))))))) ( -25.30) >DroSim_CAF1 17568 99 + 1 CGUAAAAU----------AUAUGAUUAAAAUG-GAAAAAACAACCCGCCUAUUUGUAUGUAUGUGUGUGUGAGUGACGCGCCAACUUGCAUGGCAUUUCGUACAUACAAA ........----------..............-..................((((((((((((...((((.....))))((((.......))))....)))))))))))) ( -20.90) >DroYak_CAF1 19927 110 + 1 CAUUAAAUCCUAUAAGAAAAAUGGUUUAAAGGGUGAAAAGCAACCCGCUUGCAUGUAUGUAUGUAUGUAAGGGCGACUUGCCAAUUUGCGUGGCACGUCGUACAUACAAA ..(((((.((............))))))).((((........)))).......((((((((((.((((.(((....)))((((.......)))))))))))))))))).. ( -29.70) >consensus CGUAAAAU__________AUAUGAUUAAAAGGGGAAAAAACAACCCGCCUAUUUGUAUGUAUGUAUGUGUGAGUGACGCGCCAACUUGCAUGGCAUUUCGUACAUACAAA ....................................................(((((((((((.....(((.....)))((((.......))))....))))))))))). (-18.36 = -17.43 + -0.94)

| Location | 588,496 – 588,592 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 71.79 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.61 |
| Structure conservation index | 0.74 |
| SVM decision value | 6.08 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 588496 96 + 22407834 CAACCCGCUUGUUUGUAUGCAUGCAUGUGUGUGUGACGUGCCAAUUUGCAUGGCGUUUCGUGCAUACAAACAUACAUAAAUAUGUAUGUAAGUGCA .....(((((..((((((((((((((....))))((..(((((.......)))))..))))))))))))((((((((....))))))))))))).. ( -33.10) >DroSec_CAF1 19803 76 + 1 CAACCCGCCUAUUUGUAUGUAUGUAUGUGUGAGUGACGCGCCAACUUGCAUGGCAUUUCGUACAUACAAACAUACA-------------------- .............(((((((.((((((((((((.....)((((.......))))...))))))))))).)))))))-------------------- ( -23.10) >DroYak_CAF1 19967 88 + 1 CAACCCGCUUGCAUGUAUGUAUGUAUGUAAGGGCGACUUGCCAAUUUGCGUGGCACGUCGUACAUACAAACAUACAUACAUAUU--------UGCA ......((..(.((((((((((((.((((.(..((((.(((((.......))))).))))..).)))).)))))))))))).).--------.)). ( -38.10) >consensus CAACCCGCUUGUUUGUAUGUAUGUAUGUGUGAGUGACGUGCCAAUUUGCAUGGCAUUUCGUACAUACAAACAUACAUA_AUAU_________UGCA ...............(((((((((.((((((..(((..(((((.......)))))..)))..)))))).))))))))).................. (-23.28 = -24.73 + 1.45)


| Location | 588,496 – 588,592 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 71.79 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.27 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.73 |
| SVM decision value | 6.79 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 588496 96 - 22407834 UGCACUUACAUACAUAUUUAUGUAUGUUUGUAUGCACGAAACGCCAUGCAAAUUGGCACGUCACACACACAUGCAUGCAUACAAACAAGCGGGUUG .((...((((((......))))))((((((((((((......((((.......))))..((.....)).......)))))))))))).))...... ( -26.90) >DroSec_CAF1 19803 76 - 1 --------------------UGUAUGUUUGUAUGUACGAAAUGCCAUGCAAGUUGGCGCGUCACUCACACAUACAUACAUACAAAUAGGCGGGUUG --------------------(((((((.(((((((..((..(((((.......)))))..))......))))))).)))))))............. ( -22.30) >DroYak_CAF1 19967 88 - 1 UGCA--------AAUAUGUAUGUAUGUUUGUAUGUACGACGUGCCACGCAAAUUGGCAAGUCGCCCUUACAUACAUACAUACAUGCAAGCGGGUUG .((.--------..(((((((((((((.((((.(..((((.(((((.......))))).))))..).)))).)))))))))))))...))...... ( -36.30) >consensus UGCA_________AUAU_UAUGUAUGUUUGUAUGUACGAAAUGCCAUGCAAAUUGGCACGUCACACACACAUACAUACAUACAAACAAGCGGGUUG ....................(((.((((((((((((.(....((((.......))))..((.......))...).)))))))))))).)))..... (-20.82 = -20.27 + -0.55)

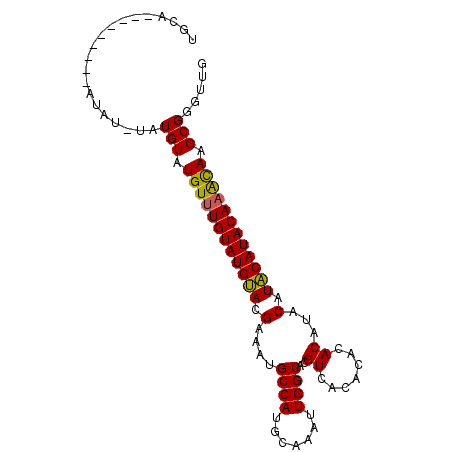
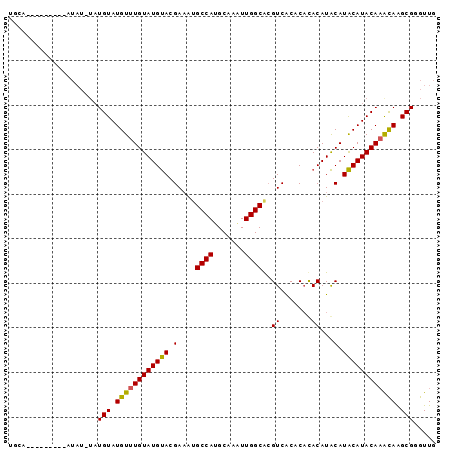
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:28:57 2006