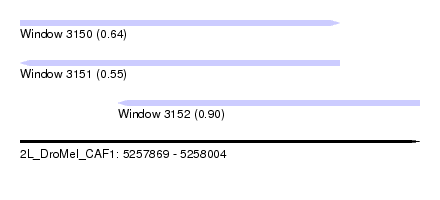
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,257,869 – 5,258,004 |
| Length | 135 |
| Max. P | 0.899139 |

| Location | 5,257,869 – 5,257,977 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -16.84 |
| Energy contribution | -18.95 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5257869 108 + 22407834 --UCUGGGCCAGCUUAUUAUAGUU-----CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUCUGGCAAUUGC-----UCUCAGUCG --..((((...(((((((((....-----.)))))))))...))))((((((....))))))....((((((.....((((........))))...))))))....-----......... ( -31.90) >DroPse_CAF1 34649 112 + 1 --------CCAUCUUAUUAAAGUCCGUUUCGUAAUGAGCAUUCCCAGCUGCAUUUCUGCAGGCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUCUGGCAAUUGGCAAUCCCUCCAUCU --------....(((((((..(......)..))))))).........(((((....)))))((((.((((((.....((((........))))...)))))).))))............. ( -25.40) >DroSec_CAF1 34667 108 + 1 --UCUGGGCCAGCUUAUUAUAGUU-----CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAACCCAUUUGCCAAUUAUAUGAAAUUAAAGUCAUAUCUGGCAAUUGC-----UCUCGCUCG --.(((((...(((((((((....-----.)))))))))...))))).((((....))))...((.((((((...((((((........)))))).)))))).)).-----......... ( -32.30) >DroYak_CAF1 36756 111 + 1 --UCUGGCCCAGCUUAUUAUAGUUGA--GCGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUCUGGCAAUUGC-----UGUCGGUCG --...((((((((.....((((..((--((((.....))))))...((((((....))))))))))((((((.....((((........))))...))))))..))-----))..)))). ( -33.60) >DroAna_CAF1 15984 108 + 1 GGCCUGGCCCAGCUUAUUAAAGUU-----CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGACCAUUAGCCAAUUAAACGAAAUUAAAGUCAUAUCGGGCAAUUGC-----CGGCGA--G ((.((((....((((((((.....-----..))))))))....))))(((((....))))).))....(((.......(((.((.......)).)))(((....))-----))))..--. ( -30.30) >DroPer_CAF1 34706 112 + 1 --------CCAUCUUAUUAAAGUCCGUUUCGUAAUGAGCAUUCCCAGCUGCAUUUCUGCAGGCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUCUGGCAAUUGGCAAUCCCUCCAUCU --------....(((((((..(......)..))))))).........(((((....)))))((((.((((((.....((((........))))...)))))).))))............. ( -25.40) >consensus __UCUGG_CCAGCUUAUUAAAGUU_____CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUCUGGCAAUUGC_____CCUCGAUCG ...((((....((((((((............))))))))....))))(((((....)))))..((.((((((.....((((........))))...)))))).))............... (-16.84 = -18.95 + 2.11)

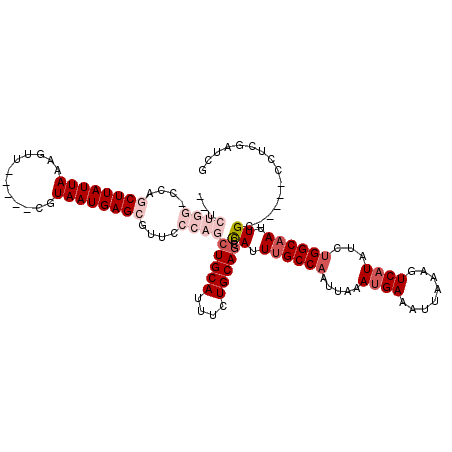
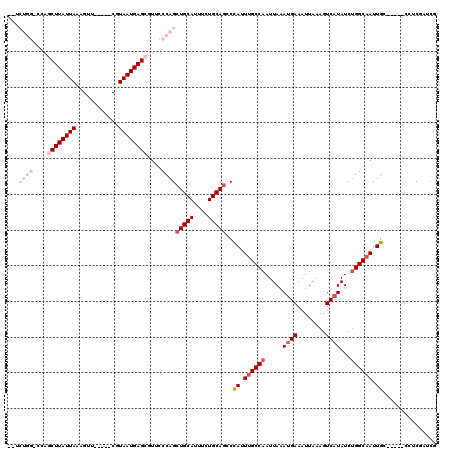
| Location | 5,257,869 – 5,257,977 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5257869 108 - 22407834 CGACUGAGA-----GCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG-----AACUAUAAUAAGCUGGCCCAGA-- .........-----....(((((((..((((........))))....)))))))...(((((((....)))))))(((...(((.((((..-----.....)))).)))...)))...-- ( -31.40) >DroPse_CAF1 34649 112 - 1 AGAUGGAGGGAUUGCCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAAUGCUCAUUACGAAACGGACUUUAAUAAGAUGG-------- ...(((((.....((((.(((((((..((((........))))....))))))).))))(((((....))))).((((....))))............))))).........-------- ( -28.60) >DroSec_CAF1 34667 108 - 1 CGAGCGAGA-----GCAAUUGCCAGAUAUGACUUUAAUUUCAUAUAAUUGGCAAAUGGGUUGCAGAAAUGCAGCUGGGAACGCUCAUUACG-----AACUAUAAUAAGCUGGCCCAGA-- ...((....-----))..((((((.((((((........))))))...))))))...(((((((....)))))))(((...(((.((((..-----.....)))).)))...)))...-- ( -35.50) >DroYak_CAF1 36756 111 - 1 CGACCGACA-----GCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACGC--UCAACUAUAAUAAGCUGGGCCAGA-- .(.((..((-----((..(((((((..((((........))))....)))))))((((((((((....))))))((((....))))......--....)))).....)))))).)...-- ( -32.10) >DroAna_CAF1 15984 108 - 1 C--UCGCCG-----GCAAUUGCCCGAUAUGACUUUAAUUUCGUUUAAUUGGCUAAUGGUCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG-----AACUUUAAUAAGCUGGGCCAGGCC .--..((((-----.((((((..(((.((.......)).)))..)))))).)...(((((((((....)))((((..(((.(.((.....)-----).))))....))))))))))))). ( -30.20) >DroPer_CAF1 34706 112 - 1 AGAUGGAGGGAUUGCCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAAUGCUCAUUACGAAACGGACUUUAAUAAGAUGG-------- ...(((((.....((((.(((((((..((((........))))....))))))).))))(((((....))))).((((....))))............))))).........-------- ( -28.60) >consensus CGAUCGAGA_____GCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG_____AACUAUAAUAAGCUGG_CCAGA__ .....(((..........(((((((..((((........))))....))))))).(((.(((((....))))))))......)))................................... (-18.68 = -19.02 + 0.33)
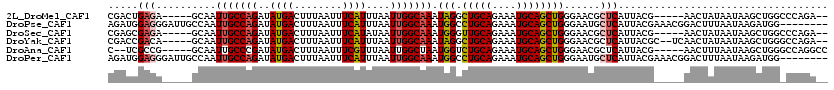
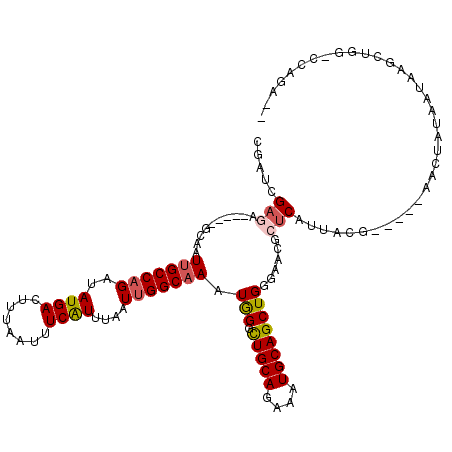
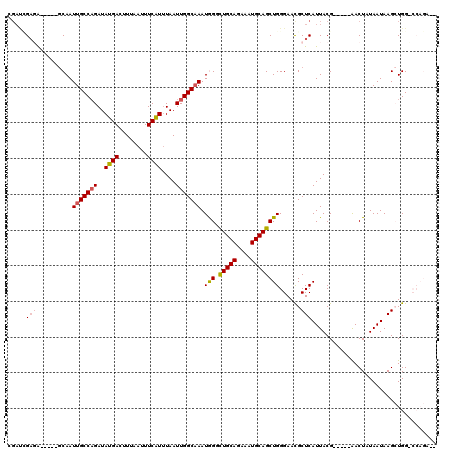
| Location | 5,257,902 – 5,258,004 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5257902 102 - 22407834 UCAUCA---GUCCCAGAAGGCAAGACGCG----------UCGACUGAGA-----GCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAA ((.(((---(((...((..((.....)).----------))))))))))-----....(((((((..((((........))))....)))))))...(((((((....)))))))..... ( -31.80) >DroPse_CAF1 34681 115 - 1 UCAUCA---GACCAAGGCGG--AGGCGCGGGCGAAGGCGGAGAUGGAGGGAUUGCCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAA ((.(((---(.(((..(((.--...)))..((....)).....))).......((((.(((((((..((((........))))....))))))).))))(((((....))))))))))). ( -37.60) >DroSec_CAF1 34700 102 - 1 UCAUCA---GUCCCAGAAGGCAAGACGCG----------UCGAGCGAGA-----GCAAUUGCCAGAUAUGACUUUAAUUUCAUAUAAUUGGCAAAUGGGUUGCAGAAAUGCAGCUGGGAA ......---.((((((.......(((.((----------(...((....-----))..((((((.((((((........))))))...))))))))).)))(((....)))..)))))). ( -34.80) >DroSim_CAF1 36194 102 - 1 UCAUCA---GUCCCAGAAGGCAAGACGCG----------UCGAGCGAGA-----GCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGGCUGCAGAAAUGCAGCUGGGAA ......---.((((.((..((.....)).----------))..((....-----))..(((((((..((((........))))....)))))))...(((((((....))))))))))). ( -34.50) >DroAna_CAF1 16019 102 - 1 UCAUCAUCUGUCCAA-ACUGGAGGCUGUG----------CC--UCGCCG-----GCAAUUGCCCGAUAUGACUUUAAUUUCGUUUAAUUGGCUAAUGGUCUGCAGAAAUGCAGCUGGGAA ..........(((..-....(((((...)----------))--))((((-----((....)))(((.((.......)).))).......)))....((.(((((....))))))).))). ( -29.50) >DroPer_CAF1 34738 115 - 1 UCAUCA---GACCAAGGCGG--AGGCGCGGGCGAAGGCGGAGAUGGAGGGAUUGCCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAA ((.(((---(.(((..(((.--...)))..((....)).....))).......((((.(((((((..((((........))))....))))))).))))(((((....))))))))))). ( -37.60) >consensus UCAUCA___GUCCAAGAAGGCAAGACGCG__________UCGAUCGAGA_____GCAAUUGCCAGAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGGCUGCAGAAAUGCAGCUGGGAA ..........(((.............................................(((((((..((((........))))....))))))).(((.(((((....))))))))))). (-18.85 = -19.10 + 0.25)

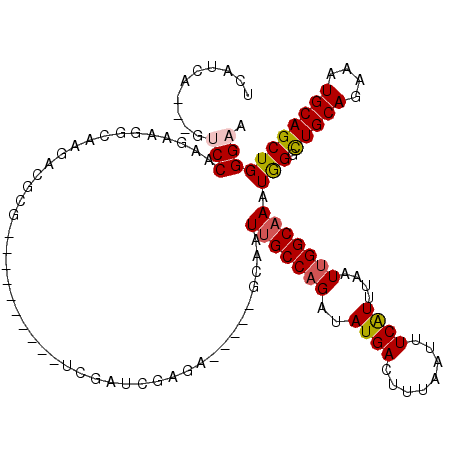
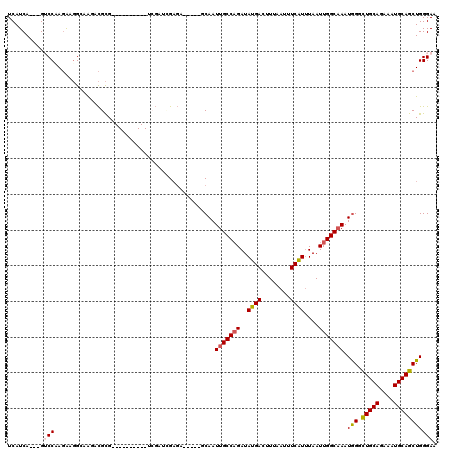
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:41 2006