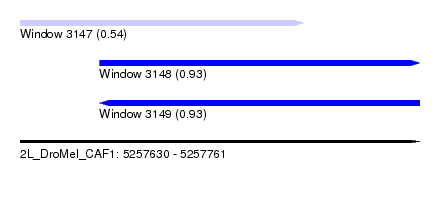
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,257,630 – 5,257,761 |
| Length | 131 |
| Max. P | 0.933596 |

| Location | 5,257,630 – 5,257,723 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -16.05 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.09 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5257630 93 + 22407834 CCAGCUC----UCUCUCUCUCGGAGGAAACUUAACAAAAUAAUAAAAAAUGAAACGAGAGGAAAAUAAAAAAUAAGAACUGCGCAUUUAAGCGCUUC .......----..((.(((((((((....)))..((.............))...))))))))..................((((......))))... ( -18.42) >DroSec_CAF1 34435 95 + 1 CCAGCUCACUCUCU--CUCUCAGAGGAAACUUAACAAAAUAAUAAAAAAUGAAACGAGAAGAAAAUAAAAAAUAAGAACUGCGCAUUUAAGCGCUUC ...........(((--.((((.(((....)))..((.............))....)))))))..................((((......))))... ( -13.72) >DroSim_CAF1 35929 97 + 1 CCAGCUCACUCUCUCUCUCUCGGAGGAAACUUAACAAAAUAAUAAAAAAUGAAACGAGAAGAAAAUAAAAAAUAAGAACUGCGCAUUUAAGCGCUUC .............(((.((((((((....)))..((.............))...))))))))..................((((......))))... ( -16.02) >consensus CCAGCUCACUCUCUCUCUCUCGGAGGAAACUUAACAAAAUAAUAAAAAAUGAAACGAGAAGAAAAUAAAAAAUAAGAACUGCGCAUUUAAGCGCUUC ...........(((...((((((((....)))..((.............))...))))))))..................((((......))))... (-12.97 = -13.09 + 0.11)


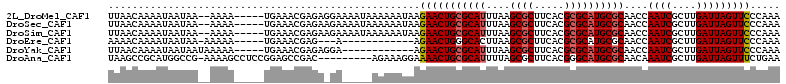
| Location | 5,257,656 – 5,257,761 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5257656 105 + 22407834 UUAACAAAAUAAUAA--AAAA-----UGAAACGAGAGGAAAAUAAAAAAUAAGAACUGCGCAUUUAAGCGCUUCACGCGCAUGCGCAACCAAUCGCUUGAUUAGUUCCCAAA ...............--....-----.....(....)...............(((((((((((....((((.....))))))))))....((((....)))))))))..... ( -20.20) >DroSec_CAF1 34463 105 + 1 UUAACAAAAUAAUAA--AAAA-----UGAAACGAGAAGAAAAUAAAAAAUAAGAACUGCGCAUUUAAGCGCUUCACGCGCAUGCGCAACCAAUCGCUUGAUUAGUUCCCAAA ...............--....-----..........................(((((((((((....((((.....))))))))))....((((....)))))))))..... ( -19.80) >DroSim_CAF1 35959 105 + 1 UUAACAAAAUAAUAA--AAAA-----UGAAACGAGAAGAAAAUAAAAAAUAAGAACUGCGCAUUUAAGCGCUUCACGCGCAUGCGCAACCAAUCGCUUGAUUAGUUCCCAAA ...............--....-----..........................(((((((((((....((((.....))))))))))....((((....)))))))))..... ( -19.80) >DroEre_CAF1 27589 91 + 1 AAAACAAAAUAAUAA-AAAAA-----UGAAACGAG---A------------AGAACUGGGCACUUAAGCGCUUCACGCGCAUGCGCAACCAAUCGCUUGAUUAGUUCCCAAA ...............-.....-----.........---.------------.....((((.(((((((((......(((....))).......))))))...))).)))).. ( -17.22) >DroYak_CAF1 36555 95 + 1 UUAACAAAAUAAUAAUAAAAA-----UGAAACGAGAGGA------------AGAACUGCGCAUUUAAGCGCUUCACGCGCAUGCGCAACCAAUCGCUUGAUUAGUUCCCAAA .....................-----..........((.------------.(((((((((((....((((.....))))))))))....((((....)))))))))))... ( -21.20) >DroAna_CAF1 15751 102 + 1 UAAGCCGCAUGGCCG-AAAAGCCUCCGGAGCCGAC---------AGAAAGGAAAACUGCGCAUUUUAGCGCUUCACGGGCAUGCGCAACAAAUCGCUUGAUUAGUUUCUGAA ...((.(((((.(((-.........((....))..---------.(((((.....))((((......))))))).))).)))))))(((.((((....)))).)))...... ( -27.20) >consensus UUAACAAAAUAAUAA__AAAA_____UGAAACGAGA_GA_____AAAAAUAAGAACUGCGCAUUUAAGCGCUUCACGCGCAUGCGCAACCAAUCGCUUGAUUAGUUCCCAAA ....................................................(((((((((((....((((.....))))))))))....((((....)))))))))..... (-17.03 = -17.25 + 0.22)
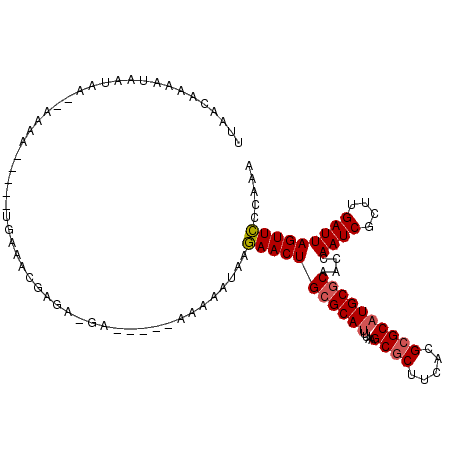
| Location | 5,257,656 – 5,257,761 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5257656 105 - 22407834 UUUGGGAACUAAUCAAGCGAUUGGUUGCGCAUGCGCGUGAAGCGCUUAAAUGCGCAGUUCUUAUUUUUUAUUUUCCUCUCGUUUCA-----UUUU--UUAUUAUUUUGUUAA ..((((((((((((((....))))))((((((((((.....))))....)))))))))))))).......................-----....--............... ( -25.90) >DroSec_CAF1 34463 105 - 1 UUUGGGAACUAAUCAAGCGAUUGGUUGCGCAUGCGCGUGAAGCGCUUAAAUGCGCAGUUCUUAUUUUUUAUUUUCUUCUCGUUUCA-----UUUU--UUAUUAUUUUGUUAA ..((((((((((((((....))))))((((((((((.....))))....)))))))))))))).......................-----....--............... ( -25.90) >DroSim_CAF1 35959 105 - 1 UUUGGGAACUAAUCAAGCGAUUGGUUGCGCAUGCGCGUGAAGCGCUUAAAUGCGCAGUUCUUAUUUUUUAUUUUCUUCUCGUUUCA-----UUUU--UUAUUAUUUUGUUAA ..((((((((((((((....))))))((((((((((.....))))....)))))))))))))).......................-----....--............... ( -25.90) >DroEre_CAF1 27589 91 - 1 UUUGGGAACUAAUCAAGCGAUUGGUUGCGCAUGCGCGUGAAGCGCUUAAGUGCCCAGUUCU------------U---CUCGUUUCA-----UUUUU-UUAUUAUUUUGUUUU .((((.((((((((....))))))))((((..((((.....))))....))))))))....------------.---.........-----.....-............... ( -22.00) >DroYak_CAF1 36555 95 - 1 UUUGGGAACUAAUCAAGCGAUUGGUUGCGCAUGCGCGUGAAGCGCUUAAAUGCGCAGUUCU------------UCCUCUCGUUUCA-----UUUUUAUUAUUAUUUUGUUAA ...(((((((((((....)))))))(((((((((((.....))))....))))))).....------------)))).........-----..................... ( -25.70) >DroAna_CAF1 15751 102 - 1 UUCAGAAACUAAUCAAGCGAUUUGUUGCGCAUGCCCGUGAAGCGCUAAAAUGCGCAGUUUUCCUUUCU---------GUCGGCUCCGGAGGCUUUU-CGGCCAUGCGGCUUA ......(((.((((....)))).))).((((((.(((.(((((.((.....((((((.........))---------))..)).....))))))).-))).))))))..... ( -29.40) >consensus UUUGGGAACUAAUCAAGCGAUUGGUUGCGCAUGCGCGUGAAGCGCUUAAAUGCGCAGUUCUUAUUUUU_____UC_UCUCGUUUCA_____UUUU__UUAUUAUUUUGUUAA ....((((((((((((....))))))((((((((((.....))))....))))))))))))................................................... (-21.10 = -21.18 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:38 2006