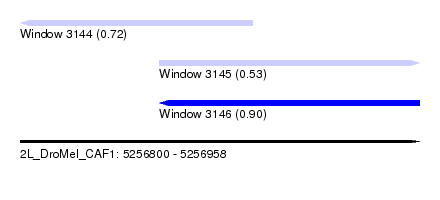
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,256,800 – 5,256,958 |
| Length | 158 |
| Max. P | 0.901470 |

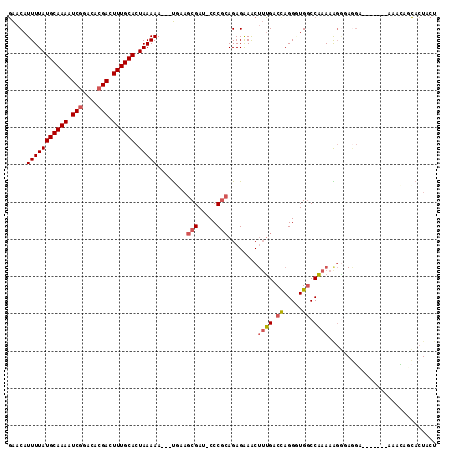
| Location | 5,256,800 – 5,256,892 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.86 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -13.87 |
| Energy contribution | -13.23 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
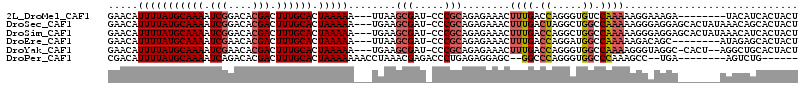
>2L_DroMel_CAF1 5256800 92 - 22407834 UAGUGCAAAGUCGUGUCCGAUUUUGCAUAAAAUGUUCG------CCGACUCCAACUGAAAUGGGAAAUGGAAGUGCAAAUGGAAAUGGCAAUGGCAAU ..(((((((((((....)))))))))))....((((.(------(((..(((((((...((.....))...))).....))))..))))...)))).. ( -24.10) >DroPse_CAF1 32524 92 - 1 UAGUGCAAAGUCGUGUCUGAUUUUGCAUAAAAUGUCGGGGAUUCUCGACUAAAAUUGAAAUUGGAAAUGGCAGUGCAAAUGGAAAUGG------AAAU ...((((..(((((.((..(((((.........((((((....)))))).......)))))..)).)))))..))))...........------.... ( -24.69) >DroSec_CAF1 33608 92 - 1 UAGUGCAAAGUCGUGUCCGAUUUUGCAUAAAAUGUUCG------CCGUCUCCAACUGAAAUGGGAAAUGGAAGUGCAAAUGGAAAUGGCAACGGCAAU ..(((((((((((....)))))))))))....((((.(------((((.(((((((...((.....))...))).....)))).)))))))))..... ( -27.50) >DroEre_CAF1 26724 92 - 1 UAGUGCAAAGUCGUGUUCGAUUUUGCAUAAAAUGUUCG------CCGACUCCAGCUGAAAUGGGAAAUGGAAGUGCAAAUGGCAAUGGAAAUGGAAAU ..(((((((((((....))))))))))).........(------(((((((((.((......))...))).))).....))))............... ( -23.20) >DroYak_CAF1 35658 80 - 1 UAGUGCAAAGUCGUGUUCGAUUUUGCAUAAAAUGUUCG------CCGACUUCAACUGAAAUGGGAAAUGGAAGUGCAAA------------UGGAAAU ..(((((((((((....))))))))))).........(------(..(((((..((.....))......)))))))...------------....... ( -18.80) >DroPer_CAF1 32611 92 - 1 UAGUGCAAAGUCGUGUCUGAUUUUGCAUAAAAUGUCGGGGAUUCUCGACUAAAAUUGAAAUUGGAAAUGGCAGUGCAAAUGAAAAUGG------AAAU ...((((..(((((.((..(((((.........((((((....)))))).......)))))..)).)))))..))))...........------.... ( -24.69) >consensus UAGUGCAAAGUCGUGUCCGAUUUUGCAUAAAAUGUUCG______CCGACUCCAACUGAAAUGGGAAAUGGAAGUGCAAAUGGAAAUGG___UGGAAAU ..(((((((((((....)))))))))))................(((..((((.......))))...)))............................ (-13.87 = -13.23 + -0.64)

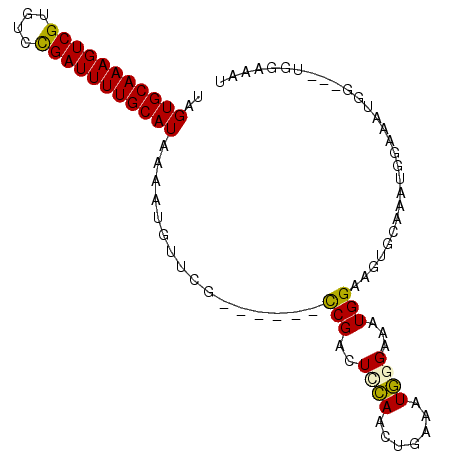
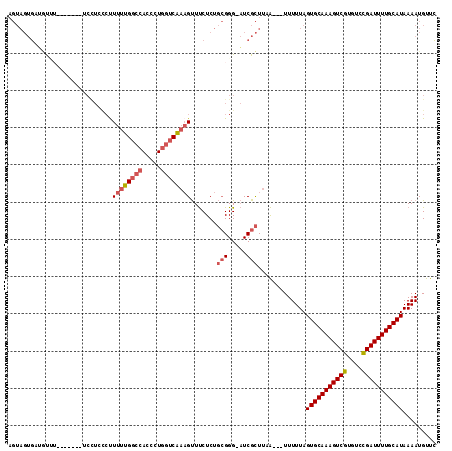
| Location | 5,256,855 – 5,256,958 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
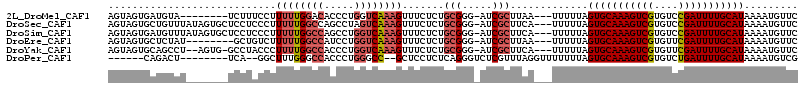
>2L_DroMel_CAF1 5256855 103 + 22407834 GAACAUUUUAUGCAAAAUCGGACACGACUUUGCACUAAAAA---UUAAGCGAU-CCCGCAGAGAAACUUUGACCAGGGUGUCCAAAAAGGAAAGA--------UACAUCACUACU .....(((((((((((.(((....))).)))))).))))).---...((.(((-(((.(((((...)))))....)))..(((.....)))....--------...))).))... ( -21.70) >DroSec_CAF1 33663 111 + 1 GAACAUUUUAUGCAAAAUCGGACACGACUUUGCACUAAAAA---UGAAGCGAU-CCCGCAGAGAAACUUUGACUAGGCUGGCCAAAAAGGGAGGAGCACUAUAAACAGCACUACU ...((((((.((((((.(((....))).))))))...))))---))..((..(-(((.(((((...)))))....((....)).....))))...)).................. ( -27.20) >DroSim_CAF1 35160 111 + 1 GAACAUUUUAUGCAAAAUCGGACACGACUUUGCACUAAAAA---UGAAGCGAU-CCCGCAGAGAAACUUUGACCAGGCUGGCCAAAAAGGGAGGAGCACUAUAAACAUCACUACU ((.((((((.((((((.(((....))).))))))...))))---))..((..(-(((.(((((...)))))....((....)).....))))...))..........))...... ( -27.30) >DroEre_CAF1 26779 103 + 1 GAACAUUUUAUGCAAAAUCGAACACGACUUUGCACUAAAAA---UUAAGCGAU-CCCGCAGAGAAACUUUGACCAGGAUGGCCAAAAAGACAGC--------AUAGAGCACUACU .....(((((((((((.(((....))).)))))).))))).---....((.((-((..(((((...)))))....)))).))..........((--------.....))...... ( -21.80) >DroYak_CAF1 35701 108 + 1 GAACAUUUUAUGCAAAAUCGAACACGACUUUGCACUAAAAA---UGAAGCGAU-CCCGCAGAGAAACUUUGACCAGGGUGGCCAAAAGGGUAGGC-CACU--AGGCUGCACUACU ...((((((.((((((.(((....))).))))))...))))---))..(((..-(((.(((((...)))))....))((((((.........)))-))).--.)..)))...... ( -30.10) >DroPer_CAF1 32666 97 + 1 CGACAUUUUAUGCAAAAUCAGACACGACUUUGCACUAAAAAAACCUAAACGAGACCCUGAGAGGAGC--GGCCCAGGGUGGCCCAAAGCC--UGA--------AGUCUG------ .(((.(((((((((((.((......)).)))))).))))).............((((((...((...--..))))))))(((.....)))--...--------.)))..------ ( -23.80) >consensus GAACAUUUUAUGCAAAAUCGGACACGACUUUGCACUAAAAA___UGAAGCGAU_CCCGCAGAGAAACUUUGACCAGGGUGGCCAAAAAGGGAGGA_______AAACAGCACUACU .....(((((((((((.(((....))).)))))).)))))........(((.....)))........((((.((.....)).))))............................. (-13.68 = -14.27 + 0.59)

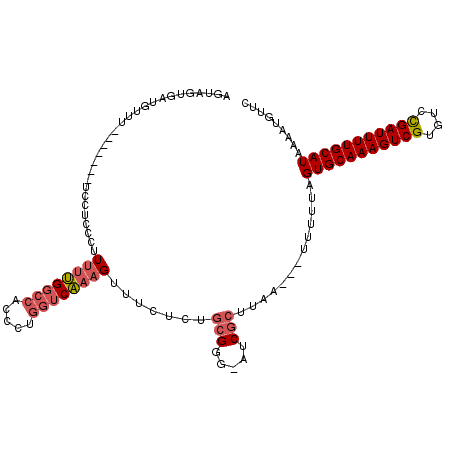
| Location | 5,256,855 – 5,256,958 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -19.55 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5256855 103 - 22407834 AGUAGUGAUGUA--------UCUUUCCUUUUUGGACACCCUGGUCAAAGUUUCUCUGCGGG-AUCGCUUAA---UUUUUAGUGCAAAGUCGUGUCCGAUUUUGCAUAAAAUGUUC ...((((((...--------....(((.....)))..((((((...........))).)))-))))))...---......(((((((((((....)))))))))))......... ( -24.30) >DroSec_CAF1 33663 111 - 1 AGUAGUGCUGUUUAUAGUGCUCCUCCCUUUUUGGCCAGCCUAGUCAAAGUUUCUCUGCGGG-AUCGCUUCA---UUUUUAGUGCAAAGUCGUGUCCGAUUUUGCAUAAAAUGUUC .(.((..(((....)))..)).)((((..((((((.......))))))((......)))))-)......((---((((..(((((((((((....)))))))))))))))))... ( -30.30) >DroSim_CAF1 35160 111 - 1 AGUAGUGAUGUUUAUAGUGCUCCUCCCUUUUUGGCCAGCCUGGUCAAAGUUUCUCUGCGGG-AUCGCUUCA---UUUUUAGUGCAAAGUCGUGUCCGAUUUUGCAUAAAAUGUUC ...(((((.((.......))...((((..(((((((.....)))))))((......)))))-)))))).((---((((..(((((((((((....)))))))))))))))))... ( -32.20) >DroEre_CAF1 26779 103 - 1 AGUAGUGCUCUAU--------GCUGUCUUUUUGGCCAUCCUGGUCAAAGUUUCUCUGCGGG-AUCGCUUAA---UUUUUAGUGCAAAGUCGUGUUCGAUUUUGCAUAAAAUGUUC ...((((.(((.(--------((.(...((((((((.....))))))))...)...)))))-).))))...---......(((((((((((....)))))))))))......... ( -30.10) >DroYak_CAF1 35701 108 - 1 AGUAGUGCAGCCU--AGUG-GCCUACCCUUUUGGCCACCCUGGUCAAAGUUUCUCUGCGGG-AUCGCUUCA---UUUUUAGUGCAAAGUCGUGUUCGAUUUUGCAUAAAAUGUUC ......((.(((.--...)-))...(((.(((((((.....)))))))((......)))))-...))..((---((((..(((((((((((....)))))))))))))))))... ( -35.20) >DroPer_CAF1 32666 97 - 1 ------CAGACU--------UCA--GGCUUUGGGCCACCCUGGGCC--GCUCCUCUCAGGGUCUCGUUUAGGUUUUUUUAGUGCAAAGUCGUGUCUGAUUUUGCAUAAAAUGUCG ------..(((.--------...--(((.....)))((((((((..--......))))))))..................(((((((((((....))))))))))).....))). ( -30.70) >consensus AGUAGUGAUGUUU_______UCCUCCCUUUUUGGCCACCCUGGUCAAAGUUUCUCUGCGGG_AUCGCUUAA___UUUUUAGUGCAAAGUCGUGUCCGAUUUUGCAUAAAAUGUUC ............................((((((((.....)))))))).......(((.....))).............(((((((((((....)))))))))))......... (-19.55 = -20.30 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:36 2006