| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,254,081 – 5,254,175 |
| Length | 94 |
| Max. P | 0.908697 |

| Location | 5,254,081 – 5,254,175 |
|---|---|
| Length | 94 |
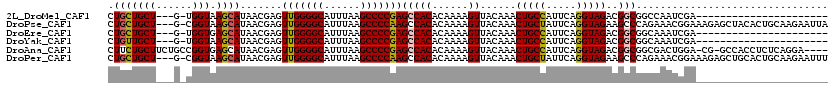
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
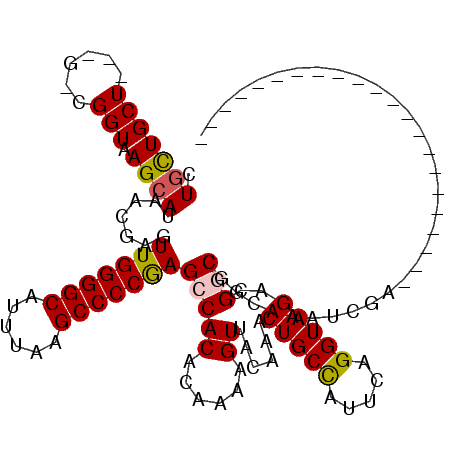
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.90 |
| Covariance contribution | -0.08 |
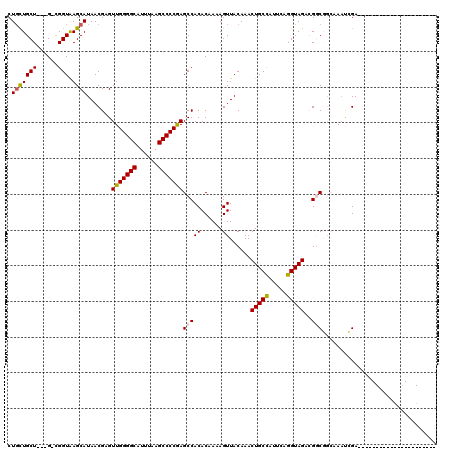
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5254081 94 - 22407834 CUGCUGCU---G-UGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCCCGAGCCACACAAAAGUUACAAACUGCCAUUCAGGUAGACGGCGGCCAAUCGA---------------------- ..((((((---(-((((.(((.......)))(((((......)))))..)))))..............(((((.....)))))..)))))).......---------------------- ( -33.50) >DroPse_CAF1 29804 116 - 1 CUGCUGCU---G-CGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCCCAAGCCACACAAAAGUUACAAACUGCUAUUCAGGUAGAAGCCCAGAAACGGAAAGAGCUACACUGCAAGAAUUA .......(---(-((((.(((.....(..(((((((......)))))))..)................(((((.....)))))....((......)).....)))..))))))....... ( -28.60) >DroEre_CAF1 23815 94 - 1 CUGCUGCU---G-UGGUGAGCAUAACGAGUUGGGGCAUUUAAGCCCCGAGCCACACAAAAGUUACAAACUGCCAUUCAGGUAGACGGCGGCAAAUCGA---------------------- .(((((((---(-(.(((.((........(((((((......))))))))))))..............(((((.....))))))))))))))......---------------------- ( -36.70) >DroYak_CAF1 32914 94 - 1 CUGUUGCU---G-UGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCCCGAGCCACACAAAAGUUACAAACUGCCAUUCAGGUAGACGGCGGCAAAUCGA---------------------- .(((((((---(-((((.(((.......)))(((((......)))))..)))))..............(((((.....)))))..)))))))......---------------------- ( -32.70) >DroAna_CAF1 12113 114 - 1 CUUCUGCUUCUGCCGGUGAGCAUAACGAGUUGGGGCAUUUAAGCCCCGAGCCACACAAAAGUUACAAACUGCCAUUCAGGUAGACGGCGGCGACUGGA-CG-GCCACCUCUCAGGA---- .......(((((..((((.((....(.(((((((((......)))))..(((((......))......(((((.....)))))..)))...)))).).-..-))))))...)))))---- ( -37.40) >DroPer_CAF1 29876 116 - 1 CUGCUGCU---G-CGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCCCAAGCCACACAAAAGUUACAAACUGCUAUUCAGGUAGAAGCCCAGAAACGGAAAGAGCUGCACUGCAAGAAUUU .(((.(.(---(-((((..((.....(..(((((((......)))))))..)................(((((.....)))))..))((......)).....))))))).)))....... ( -31.30) >consensus CUGCUGCU___G_CGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCCCGAGCCACACAAAAGUUACAAACUGCCAUUCAGGUAGACGGCGGCAAAUCGA______________________ .(((((((......))).)))).......(((((((......)))))))(((((......))......(((((.....)))))..)))................................ (-22.98 = -22.90 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:33 2006