
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,253,675 – 5,253,796 |
| Length | 121 |
| Max. P | 0.870163 |

| Location | 5,253,675 – 5,253,777 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5253675 102 + 22407834 ----CUCCAGCUACUCGUCGUUUGAACCC----------GU----CCCGAGUGCAGCCCCAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGGCAGACGUGGCUGGAAUUUUA ----.(((((((((((((((.........----------..----...(((.((.........))))).........((((.(((.....))))))))))).)).)))))))))...... ( -32.80) >DroPse_CAF1 28810 112 + 1 GCUACUCCUUCUACUC---CUUGGCGGACCUUCGGUUG-GG----GCCUGGGGUAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCUUGGCCGCCAAACGUGGCUGGAAUUUUA .....(((..((((..---.(((((((.((..((((((-((----((........))))))).(((((............)))))...)))...)))))))))..))))..)))...... ( -39.30) >DroEre_CAF1 23393 99 + 1 ----CUGCAGCUACUC---GUCUGAACCC----------GU----CUUGAGAGCAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGGCAGACGUGGCUGGAAUUUUA ----...(((((((..---(((((...((----------(.----.(((((.((.........)).)))))......((((.(((.....))))))))))))))))))))))........ ( -31.10) >DroYak_CAF1 32469 107 + 1 ---GCUCCAGCUACUC---GUUUGAAGCUCUUGAA-A--GU----CUUGAGAGCAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGACAGACGUGGCUGGAAUUUUA ---..(((((((((..---(((((..(((((..(.-.--..----.)..))))).......................((((.(((.....)))))))...))))))))))))))...... ( -39.60) >DroAna_CAF1 11710 112 + 1 ---GCUCCUGCUCCUU---GCUUGCUCCU-UUGAA-UGCUACUUUUUUUCUUCCUGCCCCAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCGGGCCGGCCGACGUGGCUGGAAUUUUA ---..(((.((...((---((((((....-(((((-.(((......................))).)))))......))..))))))...)).)))((((((.....))))))....... ( -23.55) >DroPer_CAF1 28891 112 + 1 GCUACUCCUUCUACUC---CUUGGCGGACCUUCGGUUG-GG----GCCUGGGGUAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCUUGGCCGCCAAACGUGGCUGGAAUUUUA .....(((..((((..---.(((((((.((..((((((-((----((........))))))).(((((............)))))...)))...)))))))))..))))..)))...... ( -39.30) >consensus ____CUCCAGCUACUC___GUUUGAACCC_UU____U__GU____CCCGAGAGCAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGGCAGACGUGGCUGGAAUUUUA .....(((.(((((.........................................((......))............((((.(((.....)))))))........))))).)))...... (-16.70 = -17.32 + 0.61)

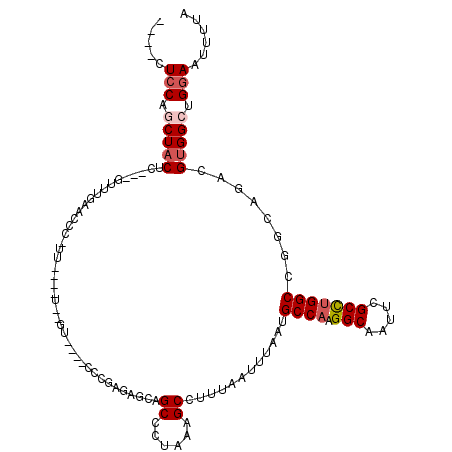

| Location | 5,253,675 – 5,253,777 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5253675 102 - 22407834 UAAAAUUCCAGCCACGUCUGCCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUGGGGCUGCACUCGGG----AC----------GGGUUCAAACGACGAGUAGCUGGAG---- ......((((((..((((.(((.(((((((.....))).))))...........)))((((((.((((......)----.)----------)).)))))).))))....)))))).---- ( -34.50) >DroPse_CAF1 28810 112 - 1 UAAAAUUCCAGCCACGUUUGGCGGCCAAGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUACCCCAGGC----CC-CAACCGAAGGUCCGCCAAG---GAGUAGAAGGAGUAGC ....(((((..(.((.((((((((((...((....(((((............)))))....(((((......)))----))-....))..)).))))))))---..)).)..)))))... ( -38.00) >DroEre_CAF1 23393 99 - 1 UAAAAUUCCAGCCACGUCUGCCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUGCUCUCAAG----AC----------GGGUUCAGAC---GAGUAGCUGCAG---- ........((((..((((((((((((((((.....))).))))...........((((....)))).........----.)----------))...)))))---)....))))...---- ( -30.30) >DroYak_CAF1 32469 107 - 1 UAAAAUUCCAGCCACGUCUGUCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUGCUCUCAAG----AC--U-UUCAAGAGCUUCAAAC---GAGUAGCUGGAGC--- .....((((((((.((.....))(((((((.....))).))))...........))))....((((((((..(((----.(--(-(....)))))).....---)))))))))))).--- ( -33.60) >DroAna_CAF1 11710 112 - 1 UAAAAUUCCAGCCACGUCGGCCGGCCCGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUGGGGCAGGAAGAAAAAAAGUAGCA-UUCAA-AGGAGCAAGC---AAGGAGCAGGAGC--- .....((((.(((.((((((.....))))))....(((((............))))).....))).))))...........((.-((...-..))))..((---.....))......--- ( -26.50) >DroPer_CAF1 28891 112 - 1 UAAAAUUCCAGCCACGUUUGGCGGCCAAGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUACCCCAGGC----CC-CAACCGAAGGUCCGCCAAG---GAGUAGAAGGAGUAGC ....(((((..(.((.((((((((((...((....(((((............)))))....(((((......)))----))-....))..)).))))))))---..)).)..)))))... ( -38.00) >consensus UAAAAUUCCAGCCACGUCUGCCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUGCACUAAAG____AC__A____AA_GGCUGCAAAC___GAGUAGCAGGAG____ ........(((((.((((((.....))))))....(((((............))))).....)))))..................................................... (-20.50 = -20.28 + -0.22)



| Location | 5,253,701 – 5,253,796 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -18.01 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5253701 95 + 22407834 U----CCCGAGUGCAGCCCCAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGGCAGACGUGGCUGGAAUUUUAAAUGUAUUUUU----CAGCCACA----------------- .----.(((((.((.........))))..........((((.(((.....)))))))))).....((((((((((.............)))----))))))).----------------- ( -29.02) >DroPse_CAF1 28846 116 + 1 G----GCCUGGGGUAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCUUGGCCGCCAAACGUGGCUGGAAUUUUAAAUGUAUUUCAACAACAGCCACAGCCACAGCCAUAGCCAC (----((.(((((..(((...((((.((......))(((....)))....)))))))..))....(((((((.......................)))))))..)))..)))........ ( -32.60) >DroEre_CAF1 23416 95 + 1 U----CUUGAGAGCAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGGCAGACGUGGCUGGAAUUUUAAAUGUAUUUUU----CAGCCACA----------------- .----..........(((.((((........))))..((((.(((.....))))))).)))....((((((((((.............)))----))))))).----------------- ( -29.82) >DroYak_CAF1 32500 95 + 1 U----CUUGAGAGCAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGACAGACGUGGCUGGAAUUUUAAAUGUAUUUUU----CAGCCACA----------------- .----.(((((.((.........)).)))))......((((.(((.....)))))))........((((((((((.............)))----))))))).----------------- ( -27.12) >DroAna_CAF1 11742 100 + 1 ACUUUUUUUCUUCCUGCCCCAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCGGGCCGGCCGACGUGGCUGGAAUUUUAAAUGUAUUUCUG---CGUCCAGA----------------- .............(((.(((...((.((......)).))...(((.....))))))((((((.....))))))........(((((....))---))).))).----------------- ( -21.30) >DroPer_CAF1 28927 116 + 1 G----GCCUGGGGUAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCUUGGCCGCCAAACGUGGCUGGAAUUUUAAAUGUAUUUCAACAACAGCCACAGCCACAGCCAUAGCCAC (----((.(((((..(((...((((.((......))(((....)))....)))))))..))....(((((((.......................)))))))..)))..)))........ ( -32.60) >consensus U____CCCGAGAGCAGCCCUAAAGCCUUUAAUUUAAUGCCAAGGCAAUUCGCCUGGCCGGCAGACGUGGCUGGAAUUUUAAAUGUAUUUCU____CAGCCACA_________________ .....................................((((.(((.....)))))))........(((((((.......................))))))).................. (-18.01 = -18.15 + 0.14)

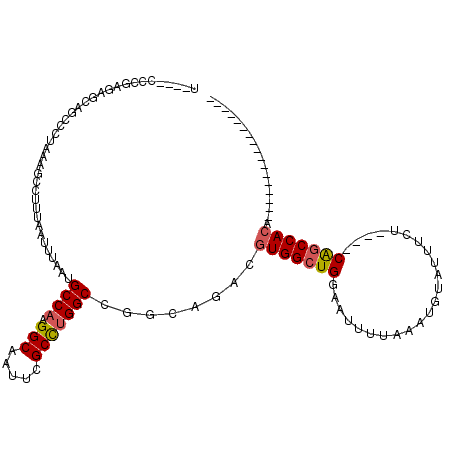
| Location | 5,253,701 – 5,253,796 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -21.36 |
| Energy contribution | -20.08 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5253701 95 - 22407834 -----------------UGUGGCUG----AAAAAUACAUUUAAAAUUCCAGCCACGUCUGCCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUGGGGCUGCACUCGGG----A -----------------.(((((((----...................))))))).....((((((((((.....))).))))...........((((....)))).....))).----. ( -29.31) >DroPse_CAF1 28846 116 - 1 GUGGCUAUGGCUGUGGCUGUGGCUGUUGUUGAAAUACAUUUAAAAUUCCAGCCACGUUUGGCGGCCAAGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUACCCCAGGC----C ...(((.(((((((.(.((((((((..(((..((.....))..)))..))))))))..).))))))))))....(((....)))..........(((((..(((....)))))))----) ( -40.50) >DroEre_CAF1 23416 95 - 1 -----------------UGUGGCUG----AAAAAUACAUUUAAAAUUCCAGCCACGUCUGCCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUGCUCUCAAG----A -----------------.(((((((----...................)))))))(((((((.(((((((.....))).))))...........)))....))))..........----. ( -27.21) >DroYak_CAF1 32500 95 - 1 -----------------UGUGGCUG----AAAAAUACAUUUAAAAUUCCAGCCACGUCUGUCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUGCUCUCAAG----A -----------------.(((((((----...................))))))).(((....(((((((.....))).))))...........((((....)))).......))----) ( -25.41) >DroAna_CAF1 11742 100 - 1 -----------------UCUGGACG---CAGAAAUACAUUUAAAAUUCCAGCCACGUCGGCCGGCCCGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUGGGGCAGGAAGAAAAAAAGU -----------------((((....---)))).............((((.(((.((((((.....))))))....(((((............))))).....))).)))).......... ( -27.00) >DroPer_CAF1 28927 116 - 1 GUGGCUAUGGCUGUGGCUGUGGCUGUUGUUGAAAUACAUUUAAAAUUCCAGCCACGUUUGGCGGCCAAGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUACCCCAGGC----C ...(((.(((((((.(.((((((((..(((..((.....))..)))..))))))))..).))))))))))....(((....)))..........(((((..(((....)))))))----) ( -40.50) >consensus _________________UGUGGCUG____AAAAAUACAUUUAAAAUUCCAGCCACGUCUGCCGGCCAGGCGAAUUGCCUUGGCAUUAAAUUAAAGGCUUUAGGGCUGCACUAAAG____A ..................((((((......(((.....)))........((((.((((((.....))))))...(((....)))..........))))....))))))............ (-21.36 = -20.08 + -1.27)

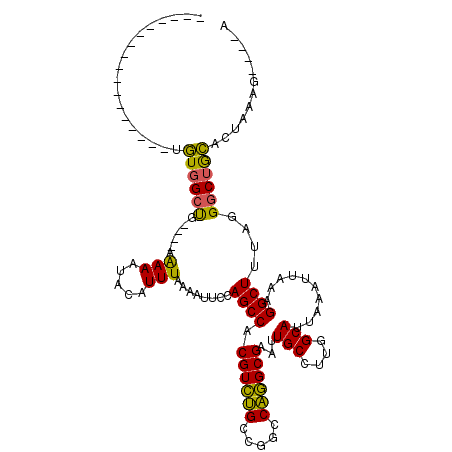
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:32 2006