| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,250,299 – 5,250,437 |
| Length | 138 |
| Max. P | 0.968172 |

| Location | 5,250,299 – 5,250,419 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
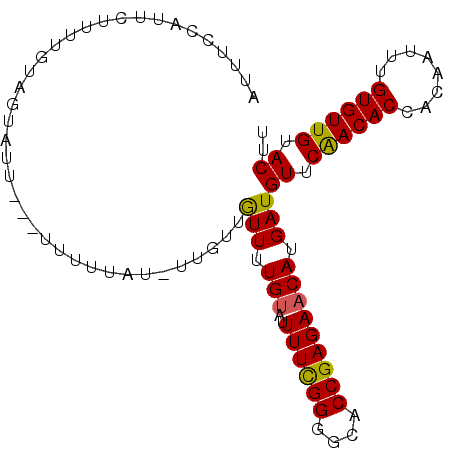
>2L_DroMel_CAF1 5250299 120 + 22407834 CAAAUCCGUACCCACUCCAUUGGCCCAUGUCGAGUACACGAAGUACAACACAAAUUGUGGUGUUGAACAUCAUGUUCUCGGUGCCCCGAAAAUACAAAACAACAAAAUAAAAAAGAAAUA .......(((((.........(((....)))(((.(((.((.((.((((((........)))))).)).)).))).)))))))).................................... ( -23.80) >DroPse_CAF1 24535 103 + 1 CAAAUCC------UCUCUGGAUCCCCAUGUCGAGUACACGAAGUACAACACAAAUUGUGGUGUCGAACAUCAUGUUCUCCUUACCCCAAAAG----AAAUAAGAC-AGAGAGA---A--- ......(------(((((((....)))((((..((((.....))))........(((.((((..((((.....))))....)))).)))...----......)))-)))))).---.--- ( -25.10) >DroEre_CAF1 20052 116 + 1 CAAAUCCGUGCCCACUCCAUAUGCCCAUGUCGAGUACACGAAGUACAACACAAAUUGUGGUGUUGAACAUCAUGUUCUCGGUGCCCCGAAAAUACAAAAUAACAA-AAAAAGA---AAUA ......((((...((((.((((....)))).)))).))))..((.((((((........)))))).))....((((.((((....)))).))))...........-.......---.... ( -23.10) >DroYak_CAF1 29361 116 + 1 CAAAUCCGUGCCCACUCCAUCUGCCCAUGUCGAGUACACGAAGUACAACACAAAUUGUGGUGUUGAACAUCAUGUUCUCGGUGCCCCGAAAAUACAAAACAACAC-ACAAAAA---AAAA ......((((...(((((((......)))..)))).))))..............(((((.((((((((.....))))((((....))))........))))...)-))))...---.... ( -21.10) >DroAna_CAF1 8344 102 + 1 CC--U--CUGCGACCUCCAAUCCUCCAUGGCGAGUACACGAAGUACAACACAAAUUGUGGUGUCGAACAUCAUGUUCUCGGUACCCCAAAA--CCAAAACAACAAAA-A----------- ..--.--....................(((.(.((((.(((.(.(((.(((.....))).))))((((.....)))))))))))))))...--..............-.----------- ( -18.20) >DroPer_CAF1 24633 103 + 1 CAAAUCC------UCUCUGGAUCCCCAUGUCGAGUACACGAAGUACAACACAAAUUGUGGUGUCGAACAUCAUGUUCUCCUUACCCCAAAAG----AAAUAAGAC-AGAGAGA---A--- ......(------(((((((....)))((((..((((.....))))........(((.((((..((((.....))))....)))).)))...----......)))-)))))).---.--- ( -25.10) >consensus CAAAUCCGUGCCCACUCCAUAUCCCCAUGUCGAGUACACGAAGUACAACACAAAUUGUGGUGUCGAACAUCAUGUUCUCGGUACCCCAAAAA_ACAAAACAACAA_AGAAAGA___A___ ..............................((((.(((.((.((.(.((((........)))).).)).)).))).))))........................................ (-12.25 = -12.58 + 0.33)


| Location | 5,250,299 – 5,250,419 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5250299 120 - 22407834 UAUUUCUUUUUUAUUUUGUUGUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCCAAUGGAGUGGGUACGGAUUUG ....(((..(((((((..(((..(..(..((((((....))))))((((((.((.((((((........)))))).)).)))))).......)..)..)))..)))))))...))).... ( -32.50) >DroPse_CAF1 24535 103 - 1 ---U---UCUCUCU-GUCUUAUUU----CUUUUGGGGUAAGGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGGAUCCAGAGA------GGAUUUG ---(---(((((((-(.(((((..----(....)..)))))(((.((((((.((.((((((........)))))).)).)))))).)))............))))))------))).... ( -38.40) >DroEre_CAF1 20052 116 - 1 UAUU---UCUUUUU-UUGUUAUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAUAUGGAGUGGGCACGGAUUUG ....---.......-.((((...........((((....))))((((.....))))))))....((((((((((.((((((((((.(((.....))).)))))))))))))))))))).. ( -30.70) >DroYak_CAF1 29361 116 - 1 UUUU---UUUUUGU-GUGUUGUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAGAUGGAGUGGGCACGGAUUUG ....---...((((-((((((..(..(.(((((((....))))))).)..)....))))).)))))((((((((.((((((((.(.(((.....))).).)))))))))))))))).... ( -36.40) >DroAna_CAF1 8344 102 - 1 -----------U-UUUUGUUGUUUUGG--UUUUGGGGUACCGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGCCAUGGAGGAUUGGAGGUCGCAG--A--GG -----------.-((((((..(((..(--((((..((...((((.((((((.((.((((((........)))))).)).)))))).))))))....)))))..)))...))))--)--). ( -29.70) >DroPer_CAF1 24633 103 - 1 ---U---UCUCUCU-GUCUUAUUU----CUUUUGGGGUAAGGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGGAUCCAGAGA------GGAUUUG ---(---(((((((-(.(((((..----(....)..)))))(((.((((((.((.((((((........)))))).)).)))))).)))............))))))------))).... ( -38.40) >consensus ___U___UCUUUCU_GUGUUAUUUUGU_UUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAUAUGGAGUGGGCACGGAUUUG ........((((...(((((((((((......))))))..((((.((((((.((.((((((........)))))).)).)))))).)))).....)))))..)))).............. (-20.26 = -20.57 + 0.31)

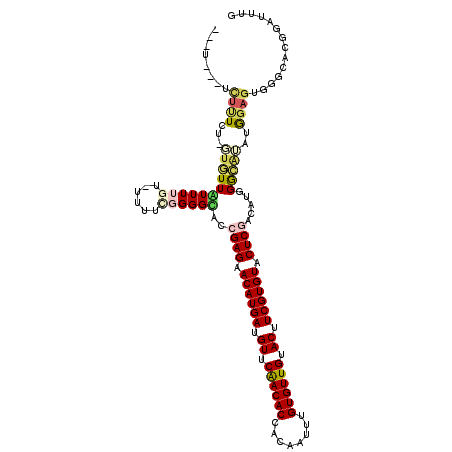
| Location | 5,250,339 – 5,250,437 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.48 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -15.13 |
| Energy contribution | -14.88 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5250339 98 - 22407834 AUUUUCAUUCUUUUGUAGUAUUUCUUUUUUAUUUUGUUGUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUU ................(((((.............(((((..(..(.(((((((....))))))).)..)....))))).(((.....)))..))))). ( -17.10) >DroSec_CAF1 27274 95 - 1 AUUUCCAUUCUUUUGUAGUAUU---UUUUUAUGUUGUUGUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCAGAAUUUGUGUUGUACUU ................(((((.---......((.(((((..(..(.(((((((....))))))).)..)....))))).))...........))))). ( -16.57) >DroSim_CAF1 28637 95 - 1 AUUUCCAUUCUUUUGUAGUAUU---UUUUUAUGUUGUUGUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUU ................(((((.---......(((((.....(..(.(((((((....))))))).)..)....))))).(((.....)))..))))). ( -17.30) >DroEre_CAF1 20092 94 - 1 AUUUUCCUUCUUUUGUAGUAUU---UCUUUUU-UUGUUAUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUU ......................---.......-........(..(.(((((((....))))))).)..).((.((((((........)))))).)).. ( -15.80) >DroYak_CAF1 29401 94 - 1 AUUUUCCUUCUUUUGUAGUUUU---UUUUUGU-GUGUUGUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUU ..............(((.....---...((((-((((((..(..(.(((((((....))))))).)..)....))))).))))).........))).. ( -19.63) >DroAna_CAF1 8380 77 - 1 CCCCCCAUUUU------------------U-UUUUGUUGUUUUGG--UUUUGGGGUACCGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUU ..(((((....------------------.-..............--...)))))......((((.....)))).(((.(((.....))).))).... ( -14.55) >consensus AUUUCCAUUCUUUUGUAGUAUU___UUUUUAU_UUGUUGUUUUGUAUUUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUU ......................................(((.(((..((((((....))))))))).)))((.((((((........)))))).)).. (-15.13 = -14.88 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:27 2006