
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,249,367 – 5,249,630 |
| Length | 263 |
| Max. P | 0.716885 |

| Location | 5,249,367 – 5,249,460 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -27.41 |
| Energy contribution | -28.77 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5249367 93 - 22407834 CUCCGUGGCCCCGCUGCCAUAACCAUGUUUUCCUCGGAAAACCCUGCUCGUUUAUCAGGCGGCAGGCAUCGGGCAA-AAGAGCUC------AGGCCGCAA ....(((((((((.((((........((((((....))))))..(((.((((.....))))))))))).)))((..-....))..------.)))))).. ( -36.50) >DroSec_CAF1 26215 93 - 1 CUCCAUGGCCCCGCUGCCAUAACCAUGUUUUCCUCGGAAAACCCUGCUCGUUUAUCAGGCGGCAGGCAUCGGGCAA-AAGAGCUC------AGGCCGCAA ......(((((((.((((........((((((....))))))..(((.((((.....))))))))))).)))((..-....))..------.)))).... ( -32.30) >DroSim_CAF1 27371 93 - 1 CUCCGUGGCCCCGCUGCCAUAACCAUGUUUUCCUCGGAAAACCCUGCUCGUUUAUCAGGCGGCAGGCAUCGGGCAA-AAGAGCUC------AGGCCGCAA ....(((((((((.((((........((((((....))))))..(((.((((.....))))))))))).)))((..-....))..------.)))))).. ( -36.50) >DroEre_CAF1 19048 93 - 1 CUCCGUGGCCCCGCUGCCAUAACCAUGUUUUCCUCGGAAAACCCUGCUCGUUUAUCAGGCGGCAGGCAUCAGGCAA-AGGAGCUC------AGGCCGCAA ....((((((((..((((........((((((....))))))(((((.((((.....))))))))).....)))).-.)).....------.)))))).. ( -36.90) >DroYak_CAF1 28370 93 - 1 CUCCGUGGCCCCGCUGCCAUAACCAUGUUUUCCUCGGAAAACCCUGCUCGUUUAUCAGGCGGCAGGCAUCGGGCAC-CAGAGCUC------AGGCCGCAA ....(((((((((.((((........((((((....))))))..(((.((((.....))))))))))).)))((..-....))..------.)))))).. ( -36.50) >DroAna_CAF1 7416 93 - 1 ------GGACCCGCAACCAUAACCAUGUUUUCCACAGAAAACUCUGUCGGUUUAUCAGGCGGCAGGCAUCGGCCAACAACAGCUCCCAGCCAGGCCGAA- ------......((............((((((....)))))).((((((.(......).)))))))).((((((.......((.....))..)))))).- ( -24.60) >consensus CUCCGUGGCCCCGCUGCCAUAACCAUGUUUUCCUCGGAAAACCCUGCUCGUUUAUCAGGCGGCAGGCAUCGGGCAA_AAGAGCUC______AGGCCGCAA ....(((((((((.((((........((((((....))))))..(((.((((.....))))))))))).)))((.......)).........)))))).. (-27.41 = -28.77 + 1.36)



| Location | 5,249,395 – 5,249,498 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.78 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5249395 103 + 22407834 CUGCCGCCUGAUAAACGAG---------------CAGGGUUUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCACGGAGGCCAGCUAUG--AGGUGGAAAGGUGCCACAAAGUGGUGGG ((.((((((.(((..((((---------------(...((((((....))))))...))).))...((..((((.....)))).))))).--))))))...))..((((......)))). ( -36.70) >DroPse_CAF1 23717 118 + 1 CCGCCGCCUGAUAGCGGGGGACGCCGUGGCAUGGCCCGACUUUCCGAGGAAAACAUGGUUAUGGCCUGGGGGCCACAGAGGCCAGCUAUGAGAGGUGGA-AGGUGGCAUAUCACAGUGG- ((.((((......)))).)).(((.(((..(((.((..((((((((.......((((((..((((((..(.....)..)))))))))))).....))))-)))))))))..))).))).- ( -49.00) >DroSec_CAF1 26243 103 + 1 CUGCCGCCUGAUAAACGAG---------------CAGGGUUUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCAUGGAGGCCAGCUAUG--AGGUGGAAAGGUGCCACAAAGUGGUGGG ((.((((((.(((..((((---------------(...((((((....))))))...))).))...((..((((.....)))).))))).--))))))...))..((((......)))). ( -36.70) >DroYak_CAF1 28398 103 + 1 CUGCCGCCUGAUAAACGAG---------------CAGGGUUUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCACGGAGGCCAGCUAUG--AGGUGGAAAGGUGCCACAAAGUGGUGGG ((.((((((.(((..((((---------------(...((((((....))))))...))).))...((..((((.....)))).))))).--))))))...))..((((......)))). ( -36.70) >DroAna_CAF1 7450 95 + 1 CUGCCGCCUGAUAAACCGA---------------CAGAGUUUUCUGUGGAAAACAUGGUUAUGGUUGCGGGUCC-------CCAGGUGGA--AGGUGGU-AGGUGCCACAAAGCAGCGGG ((((((((((.....(((.---------------(((......(((((.....)))))......))))))....-------.))))))..--..(((((-....)))))......)))). ( -31.40) >DroPer_CAF1 23808 118 + 1 CCGCCGCCUGAUAGCGGGGGACGCCGUGGCAUGGCCCGACUUUCCGAGGAAAACAUGGUUAUGGCCUGGGGGCCACAGAGGCCAGCUAUGGGAGGUGGA-AGGUGGCAUAUCACAGUGG- ..(((((((..((.(.(....).(((((((.((((((...((((....)))).........(((((....)))))..).))))))))))))..).))..-)))))))............- ( -50.10) >consensus CUGCCGCCUGAUAAACGAG_______________CAGGGUUUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCACGGAGGCCAGCUAUG__AGGUGGA_AGGUGCCACAAAGCAGUGGG ((.((((((.............................((((((....)))))).....((((((.....((((.....)))).))))))..))))))...))..((((......)))). (-23.64 = -24.78 + 1.14)


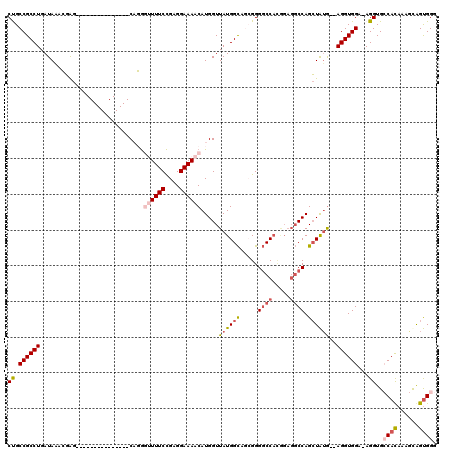
| Location | 5,249,420 – 5,249,528 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -24.71 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5249420 108 + 22407834 UUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCACGGAGGCCAGCUAUG--AGGUGGAAAGGUGCCACAAAGUGGUGGGAGUGCGACUGG----UUGUGGGAGUCGUUGCCAC ((((....)))).........(((((((((..((.((.(..((.(((((.--..((((.......))))...))))).))..).))((...----..)).))..))))))))). ( -36.30) >DroPse_CAF1 23757 97 + 1 UUUCCGAGGAAAACAUGGUUAUGGCCUGGGGGCCACAGAGGCCAGCUAUGAGAGGUGGA-AGGUGGCAUAUCACAGUGG----------------GUGUGGGAGUCAUGGUGAG ..(((.(((....(((....))).))).)))(((((.....(((.((.....)).))).-..)))))...((((.((((----------------.(.....).)))).)))). ( -30.20) >DroSec_CAF1 26268 108 + 1 UUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCAUGGAGGCCAGCUAUG--AGGUGGAAAGGUGCCACAAAGUGGUGGGAGUGCGACUGG----UUGUGGGAGUCGUUGCCAC ((((....)))).........(((((((((((((.....))))..(((..--((((.(...(.(.((((......)))).).).).)))..----)..)))...))))))))). ( -35.90) >DroEre_CAF1 19101 108 + 1 UUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCACGGAGGCCAGCUAUG--AGGUGGAAAGGUGCCACAAAGUGGUGGGAGUGCGACUGG----UUGUGGGAGUCGUUGCCUA ((((....))))..........((((((((..((.((.(..((.(((((.--..((((.......))))...))))).))..).))((...----..)).))..)))))))).. ( -35.10) >DroYak_CAF1 28423 112 + 1 UUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCACGGAGGCCAGCUAUG--AGGUGGAAAGGUGCCACAAAGUGGUGGGAGUGCGACUGGUUGGUUGUGGGAGUCGUUGCCAC ((((....)))).........(((((((((..((.((..((((((((...--.(((.(...(.(.((((......)))).).).).))))))))))).))))..))))))))). ( -40.00) >DroPer_CAF1 23848 97 + 1 UUUCCGAGGAAAACAUGGUUAUGGCCUGGGGGCCACAGAGGCCAGCUAUGGGAGGUGGA-AGGUGGCAUAUCACAGUGG----------------GUGUGGGAGUCAUGGUGAG ((((((.......((((((..((((((..(.....)..)))))))))))).....))))-))(((((...(((((....----------------.)))))..)))))...... ( -31.00) >consensus UUUCCGAGGAAAACAUGGUUAUGGCAGCGGGGCCACGGAGGCCAGCUAUG__AGGUGGAAAGGUGCCACAAAGUGGUGGGAGUGCGACUGG____UUGUGGGAGUCGUUGCCAC ((((....)))).........(((((((((..(((((....(((.((.....)).))).......((((......)))).................)))))...))))))))). (-24.71 = -24.97 + 0.25)



| Location | 5,249,528 – 5,249,630 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.82 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -24.10 |
| Energy contribution | -23.74 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5249528 102 + 22407834 AAUGUGGGCGUGCCGCACAACGCUUAGAUGUGGCCAACAAAAACGGGCAGCAAAAGCGCGUGGAGCGUGCCUCAUAAUUGGCAAC--GUGA---AUGGCCAAAGACG .((.(((((((........))))))).)).((((((......(((.((.......)).)))...(((((((........))).))--))..---.))))))...... ( -34.90) >DroPse_CAF1 23854 88 + 1 AAUGUGGGCGUGCUGCA-ACAUUU------UGGCCAGGAAA-----------GAGGCGCGUGGAGCGUGCCUCAUAAUUGGCAAAAAGUGG-AAGAGGCUGGCGACG ...((.(.((.(((...-.(((((------(.(((((....-----------((((((((.....))))))))....)))))..)))))).-....))))).).)). ( -35.80) >DroSec_CAF1 26376 102 + 1 AAUGUGGGCGUGCCGCA-AACGCUUAGAUGUGGCCAACAAAAACGGGCAGCAAAAGCGCGUGGAGCGUGCCUCAUAAUUGGCAAC--GUGAA--ACGGCCAAAGACG .((.(((((((......-.))))))).)).(((((.......(((.((.......)).)))...(((((((........))).))--))...--..)))))...... ( -33.90) >DroSim_CAF1 27532 102 + 1 AAUGUGGGCGUGCCGCA-AACGCUUAGAUGUGGCCAACAAAAACGGGCAGCAAAAGCGCGUGGAGCGUGCCUCAUAAUUGGCAAC--GUGAA--ACGGCCAAAGACG .((.(((((((......-.))))))).)).(((((.......(((.((.......)).)))...(((((((........))).))--))...--..)))))...... ( -33.90) >DroEre_CAF1 19209 100 + 1 AAUGUGGGCGUGCCACA-ACCGCUUAGAUGUGGCCAACAA-AACGGGCAGCAAAAGCGCGUGGAGCGUGCCUCAUAAUUGGCAAC--GUGA---ACGGCCAAAGACG ..(((((((((((..((-..(((((...(((.(((.....-....))).))).)))))..))..))))))).)))).(((((..(--(...---.)))))))..... ( -35.00) >DroYak_CAF1 28535 100 + 1 AAUGUGGGCGUGCCGCA-ACCGCUUAGAUGUGGCAAACAA-AACGGGCAGAAAAAGCGCGUGGAGCGUGCCUCAUAAUUGGCAAC--GUGA---AUGGCCAAAGACG ......(((.(((((((-.(......).)))))))..((.-.(((.((.......)).)))...(((((((........))).))--))..---.)))))....... ( -30.00) >consensus AAUGUGGGCGUGCCGCA_AACGCUUAGAUGUGGCCAACAAAAACGGGCAGCAAAAGCGCGUGGAGCGUGCCUCAUAAUUGGCAAC__GUGA___ACGGCCAAAGACG ..(((((((..((((((...........)))))).....................((((.....))))))).)))).(((((...............)))))..... (-24.10 = -23.74 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:24 2006