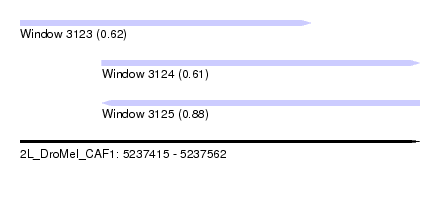
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,237,415 – 5,237,562 |
| Length | 147 |
| Max. P | 0.884756 |

| Location | 5,237,415 – 5,237,522 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.86 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5237415 107 + 22407834 CUGCUCUCGCAAAGAGAG----AGAGAGUAGACGGGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAAAAUG---CUGGCAGGCGUUUAAAAUCGCGCAACUCACACACUUGC ((((((((.(.......)----.))))))))............((((.((((((((.....)))....(((((---((....))))))).....)))))..))))......... ( -32.40) >DroSim_CAF1 15578 111 + 1 CUGCUCUCGCAAAGAGAGAGAGAGAGAGUAGUAAGGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAAAACG---CUAGCAGGCGUUUAAAAUCGCGCAACUCACACACUUGC ((((((((...............))))))))............((((.((((((((.....)))....(((((---((....))))))).....)))))..))))......... ( -32.96) >DroEre_CAF1 7347 97 + 1 CUGCUCUCGCAG-----------------AGAAAGGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAGUGCGCUGCUGGCAGGCGUUUAAAAUCGCGCCACUCACACAGUUGC ..(((((((...-----------------..............)))))))((((((.....)))..((((..((.....)).(((((........))))))))).)))...... ( -30.43) >DroYak_CAF1 16696 108 + 1 CCGCUCUCACGAAGAGA------GAGAGUAGAAAGGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAGUGAGCUGAUAGCGGGCGUUUAAAAUCGCGCAACUCACACAUUUGC ..((((((((.....).------...........(......).)))))))((((((.....)))..((((..((.....))(.(((........))).).)))).)))...... ( -28.40) >consensus CUGCUCUCGCAAAGAGA______GAGAGUAGAAAGGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAAAACG___CUAGCAGGCGUUUAAAAUCGCGCAACUCACACACUUGC ..(((((((..................................)))))))((((((.....)))(((.....((.....))..((((........))))..))).)))...... (-22.17 = -22.48 + 0.31)

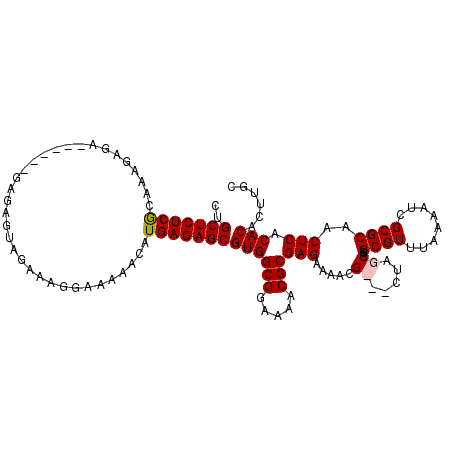
| Location | 5,237,445 – 5,237,562 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -26.64 |
| Energy contribution | -26.68 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5237445 117 + 22407834 GGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAAAAUG---CUGGCAGGCGUUUAAAAUCGCGCAACUCACACACUUGCUGGCGUUUAAAAACGCGCGUGAGGUAAAAAUACUCUCUCU ..........((((((((((((.....)))....(((((---(..(((((.((...................)).)))))..)))))).....)))))..(((.........))))))). ( -35.11) >DroSec_CAF1 14725 117 + 1 GGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAAAACG---CUAGCAGGCGUUUAAAAUCGCGCAACUCACACACUUGCUGGCGUUUAAAAACGCGCGUGAGGUAGAGAUACUCUCUCU ..........((((((((((((.....)))....(((((---((((((((.((...................)).))))))))))))).....)))))..(((.........))))))). ( -37.91) >DroSim_CAF1 15612 117 + 1 GGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAAAACG---CUAGCAGGCGUUUAAAAUCGCGCAACUCACACACUUGCUGGCGUUUAAAAACGCGCGUGAGGUAGAGAUACUCUCUCU ..........((((((((((((.....)))....(((((---((((((((.((...................)).))))))))))))).....)))))..(((.........))))))). ( -37.91) >DroEre_CAF1 7364 120 + 1 GGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAGUGCGCUGCUGGCAGGCGUUUAAAAUCGCGCCACUCACACAGUUGCUAGCGUUUAAAAACGCGCGUAAGGUAAAGAUACUCUGUCU ......(((.(((.((((........))))......((((.((((((((((((........)))))(((.....))))))))))(((....)))))))..............)))))).. ( -35.70) >DroYak_CAF1 16724 120 + 1 GGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAGUGAGCUGAUAGCGGGCGUUUAAAAUCGCGCAACUCACACAUUUGCUAGCGUUUAAAAACGCGCGUAAGGUAAAUUUACUCUCUCU .......((((....))))(((.....)))(((((((((...((.(((.((((((.....(((((((.........))))..))).....)))))).)))...))...)))))))))... ( -35.50) >consensus GGAAAAACAUGAGAGCGUGGCGGAAAACGCGAGAAAACG___CUAGCAGGCGUUUAAAAUCGCGCAACUCACACACUUGCUGGCGUUUAAAAACGCGCGUGAGGUAAAGAUACUCUCUCU ..........(((((.((((((.....)))(((.....(.......)..((((........))))..))).))).(((((..(((((....)))))..))))).........)))))... (-26.64 = -26.68 + 0.04)

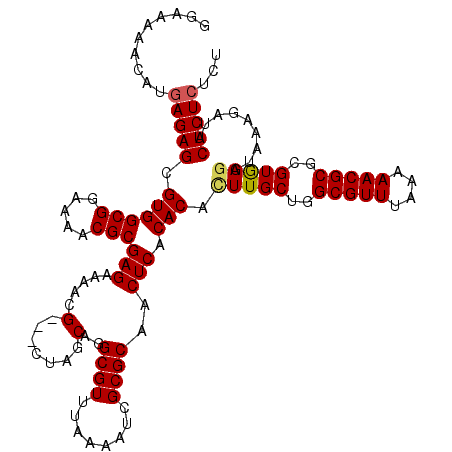
| Location | 5,237,445 – 5,237,562 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -27.41 |
| Energy contribution | -27.37 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5237445 117 - 22407834 AGAGAGAGUAUUUUUACCUCACGCGCGUUUUUAAACGCCAGCAAGUGUGUGAGUUGCGCGAUUUUAAACGCCUGCCAG---CAUUUUCUCGCGUUUUCCGCCACGCUCUCAUGUUUUUCC ...((((((.............(((((((....)))))..))..(((.(((.(..((((((....(((.((......)---).)))..))))))..).)))))))))))).......... ( -32.30) >DroSec_CAF1 14725 117 - 1 AGAGAGAGUAUCUCUACCUCACGCGCGUUUUUAAACGCCAGCAAGUGUGUGAGUUGCGCGAUUUUAAACGCCUGCUAG---CGUUUUCUCGCGUUUUCCGCCACGCUCUCAUGUUUUUCC ...((((((.............(((((((....)))))..))..(((.(((.(..((((((....((((((......)---)))))..))))))..).)))))))))))).......... ( -38.00) >DroSim_CAF1 15612 117 - 1 AGAGAGAGUAUCUCUACCUCACGCGCGUUUUUAAACGCCAGCAAGUGUGUGAGUUGCGCGAUUUUAAACGCCUGCUAG---CGUUUUCUCGCGUUUUCCGCCACGCUCUCAUGUUUUUCC ...((((((.............(((((((....)))))..))..(((.(((.(..((((((....((((((......)---)))))..))))))..).)))))))))))).......... ( -38.00) >DroEre_CAF1 7364 120 - 1 AGACAGAGUAUCUUUACCUUACGCGCGUUUUUAAACGCUAGCAACUGUGUGAGUGGCGCGAUUUUAAACGCCUGCCAGCAGCGCACUCUCGCGUUUUCCGCCACGCUCUCAUGUUUUUCC ((((((((..............(((((((....)))))..))....(((((.(((((((((.......(((.((....))))).....)))))....))))))))).))).))))).... ( -32.60) >DroYak_CAF1 16724 120 - 1 AGAGAGAGUAAAUUUACCUUACGCGCGUUUUUAAACGCUAGCAAAUGUGUGAGUUGCGCGAUUUUAAACGCCCGCUAUCAGCUCACUCUCGCGUUUUCCGCCACGCUCUCAUGUUUUUCC ...((((((...........(((((((((....)))))........(.((((((((.(((............)))...)))))))).)..))))..........)))))).......... ( -32.80) >consensus AGAGAGAGUAUCUUUACCUCACGCGCGUUUUUAAACGCCAGCAAGUGUGUGAGUUGCGCGAUUUUAAACGCCUGCUAG___CGUUUUCUCGCGUUUUCCGCCACGCUCUCAUGUUUUUCC ...((((((........((((((((((((....)))))........)))))))..(((.((....((((((...................)))))))))))...)))))).......... (-27.41 = -27.37 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:16 2006