
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,234,816 – 5,234,912 |
| Length | 96 |
| Max. P | 0.998205 |

| Location | 5,234,816 – 5,234,912 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5234816 96 + 22407834 U--------AAGCCCAAAUCAAAUGCUCAGAUGCAUAUCAAAAUAAUCUAAGCAAAUGAAAUGACAACAAAGUUGGGCAC----AC--AAAAAGGCAUAAAAUAUGUGCG .--------..((((((.(((..((((.((((.............)))).))))..)))..((....))...))))))..----..--......(((((.....))))). ( -21.72) >DroSec_CAF1 12054 100 + 1 U--------AAGCCCAAAUCAAAUGCUCAGAUGCAUAUCAAAAUAAUCUAAGCAAAUGAAAUGACAACAAAGUUGGGCACACACAC--AAAAAGGCAUAAAAUAUGUGCG .--------..((((((.(((..((((.((((.............)))).))))..)))..((....))...))))))........--......(((((.....))))). ( -21.72) >DroSim_CAF1 12940 100 + 1 U--------AAGCCCAAAUCAAAUGCUCAGAUGCAUAUCAAAAUAAUCUAAGCAAAUGAAAUGACAACAAAGUUGGGCACACACAC--AAAAAGGCAUAAAAUAUGUGCG .--------..((((((.(((..((((.((((.............)))).))))..)))..((....))...))))))........--......(((((.....))))). ( -21.72) >DroEre_CAF1 4717 96 + 1 U--------AGGCCCAAAUCAAAUGCUCAGAUGCAUAUCAAAAUAAUCUAAGCAAAUGAAAUGACAACAAAGUUGGGCAC----AC--AAAAAGACAUAAAAUAUGUGCG .--------..((((((.(((..((((.((((.............)))).))))..)))..((....))...))))))..----..--......(((((...)))))... ( -18.32) >DroYak_CAF1 13999 96 + 1 U--------AAGCCCAAAUCAAAUGCUCAGAUGCAUAUCAAAAUAAUCUAAGCAAAUGAAAUGACAACAAAGUUGGGCAC----AC--AAAAGGGCAUAAAAUAUGUGCG .--------..((((((.(((..((((.((((.............)))).))))..)))..((....))...))))))..----..--......(((((.....))))). ( -21.72) >DroPer_CAF1 3296 110 + 1 CCAGGUCCCAAAAUCAAAUCAAAUGCUCAGAUGCAUAUCAAAAUAAUCUAAGCAAAUGAAAUGACAACAAUGUUGGGCUCACACACAAAAAAAGGCAUAAAAUAUGGGCA ....((((.....(((..(((..((((.((((.............)))).))))..)))..))).....(((((..(((..............)))....))))))))). ( -17.26) >consensus U________AAGCCCAAAUCAAAUGCUCAGAUGCAUAUCAAAAUAAUCUAAGCAAAUGAAAUGACAACAAAGUUGGGCAC____AC__AAAAAGGCAUAAAAUAUGUGCG ...........((((...(((..((((.((((.............)))).))))..)))...(((......)))))))................(((((.....))))). (-17.88 = -18.40 + 0.53)

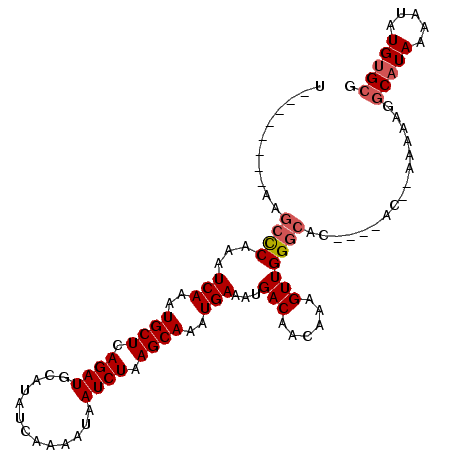
| Location | 5,234,816 – 5,234,912 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.44 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5234816 96 - 22407834 CGCACAUAUUUUAUGCCUUUUU--GU----GUGCCCAACUUUGUUGUCAUUUCAUUUGCUUAGAUUAUUUUGAUAUGCAUCUGAGCAUUUGAUUUGGGCUU--------A .(((((...............)--))----))((((((...((....))..(((..((((((((((((......))).)))))))))..))).))))))..--------. ( -26.36) >DroSec_CAF1 12054 100 - 1 CGCACAUAUUUUAUGCCUUUUU--GUGUGUGUGCCCAACUUUGUUGUCAUUUCAUUUGCUUAGAUUAUUUUGAUAUGCAUCUGAGCAUUUGAUUUGGGCUU--------A ((((((((.............)--))))))).((((((...((....))..(((..((((((((((((......))).)))))))))..))).))))))..--------. ( -30.42) >DroSim_CAF1 12940 100 - 1 CGCACAUAUUUUAUGCCUUUUU--GUGUGUGUGCCCAACUUUGUUGUCAUUUCAUUUGCUUAGAUUAUUUUGAUAUGCAUCUGAGCAUUUGAUUUGGGCUU--------A ((((((((.............)--))))))).((((((...((....))..(((..((((((((((((......))).)))))))))..))).))))))..--------. ( -30.42) >DroEre_CAF1 4717 96 - 1 CGCACAUAUUUUAUGUCUUUUU--GU----GUGCCCAACUUUGUUGUCAUUUCAUUUGCUUAGAUUAUUUUGAUAUGCAUCUGAGCAUUUGAUUUGGGCCU--------A .(((((...............)--))----))((((((...((....))..(((..((((((((((((......))).)))))))))..))).))))))..--------. ( -25.96) >DroYak_CAF1 13999 96 - 1 CGCACAUAUUUUAUGCCCUUUU--GU----GUGCCCAACUUUGUUGUCAUUUCAUUUGCUUAGAUUAUUUUGAUAUGCAUCUGAGCAUUUGAUUUGGGCUU--------A .(((((...............)--))----))((((((...((....))..(((..((((((((((((......))).)))))))))..))).))))))..--------. ( -26.36) >DroPer_CAF1 3296 110 - 1 UGCCCAUAUUUUAUGCCUUUUUUUGUGUGUGAGCCCAACAUUGUUGUCAUUUCAUUUGCUUAGAUUAUUUUGAUAUGCAUCUGAGCAUUUGAUUUGAUUUUGGGACCUGG ...(((...((((((((.......).)))))))(((((.......((((..(((..((((((((((((......))).)))))))))..)))..)))).)))))...))) ( -28.20) >consensus CGCACAUAUUUUAUGCCUUUUU__GU____GUGCCCAACUUUGUUGUCAUUUCAUUUGCUUAGAUUAUUUUGAUAUGCAUCUGAGCAUUUGAUUUGGGCUU________A ................................((((((...((....))..(((..((((((((((((......))).)))))))))..))).))))))........... (-20.66 = -20.85 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:10 2006