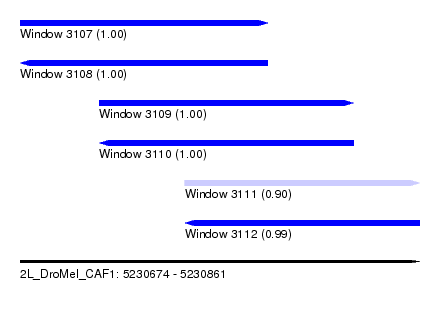
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,230,674 – 5,230,861 |
| Length | 187 |
| Max. P | 0.998790 |

| Location | 5,230,674 – 5,230,790 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -30.89 |
| Energy contribution | -32.04 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.13 |
| Structure conservation index | 0.75 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
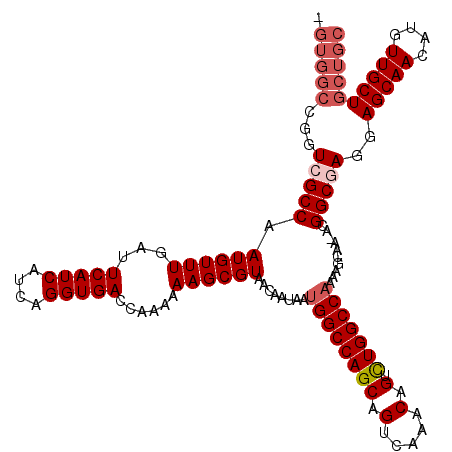
>2L_DroMel_CAF1 5230674 116 + 22407834 AAAA-AGUUCGAAAAGCAAUUCCAAACCGAUCAA--CCCAGCAACAUGUUGUUGCUGCUGCUACCGAAGCAUUUGCUACUGCAGCAGCAACAUGUUGCUUCUAGCCGU-UUGAUUUUGGC ..((-(((.((((..((...((......))....--...(((((((((((((((((((((((.....)))).........)))))))))))))))))))....))..)-))))))))... ( -39.20) >DroSec_CAF1 8026 103 + 1 AAAA-AGGUCGAAAAGCAAUUCCAAACCGAUCCGUCGCCAGCAACAUGUUGUUGC---------------AAUUGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGU-UUGCUUUUGGC ....-..(((.((((((((........(((....)))..((((((((((((((((---------------...(((....))))))))))))))))))).........-))))))))))) ( -38.20) >DroSim_CAF1 8946 103 + 1 AAAA-AGUUCGAAAAGCAAUUCCAAACCGAACC---GCCAGCAACAUGUUGUUGC------------AGCAAUUGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGU-UUGCUUUUGGC ....-................(((((.(((((.---((.((((((((((((((((------------.(((........))).))))))))))))))))....)).))-)))..))))). ( -38.60) >DroEre_CAF1 822 117 + 1 AAAAGAGAUCCAAAAGCAAUUCCAAAACGAACC---GCCAGCAACAUGUUGUUGCUGCUACAAGCGAAGCAAUUGCUACUGCAGCAGCAACAUGUUGCUCCUCGCCUUUUUGCUUUGGGC ........((((((.(((((((......)))..---((.(((((((((((((((((((....(((((.....)))))...)))))))))))))))))))....))....)))))))))). ( -46.60) >DroYak_CAF1 9826 115 + 1 AAAA-AGAUCCAAAAGCAAUUCCAAAUCGAUUC---ACCAGCAACAUGUUGUUGCCGCUACUACCGAAGCAAUUGCUACUGCAGCAGCAACAUGUUGCUCCUUGCCGU-UUGCUUUUGGC ....-....((((((((((...(((...(....---..)((((((((((((((((.(((........)))...(((....)))))))))))))))))))..)))....-)))))))))). ( -42.20) >consensus AAAA_AGAUCGAAAAGCAAUUCCAAACCGAUCC___GCCAGCAACAUGUUGUUGC_GCU_C_A_CGAAGCAAUUGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGU_UUGCUUUUGGC .........(.((((((((.((......)).........((((((((((((((((.(((........)))...(((....)))))))))))))))))))..........)))))))).). (-30.89 = -32.04 + 1.15)


| Location | 5,230,674 – 5,230,790 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -43.84 |
| Consensus MFE | -27.19 |
| Energy contribution | -28.71 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.34 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5230674 116 - 22407834 GCCAAAAUCAA-ACGGCUAGAAGCAACAUGUUGCUGCUGCAGUAGCAAAUGCUUCGGUAGCAGCAGCAACAACAUGUUGCUGGG--UUGAUCGGUUUGGAAUUGCUUUUCGAACU-UUUU ...........-.(((((...(((((((((((((((((((.(.(((....))).)....)))))))))...)))))))))).))--)))...(((((((((.....)))))))))-.... ( -46.30) >DroSec_CAF1 8026 103 - 1 GCCAAAAGCAA-ACGGCGAGUAGCAACAUGUUGCUGCUGCAGUAGCAAUU---------------GCAACAACAUGUUGCUGGCGACGGAUCGGUUUGGAAUUGCUUUUCGACCU-UUUU .(.((((((((-.((.((..(((((((((((((.((((((....)))...---------------))).))))))))))))).)).))..((......)).)))))))).)....-.... ( -35.50) >DroSim_CAF1 8946 103 - 1 GCCAAAAGCAA-ACGGCGAGUAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCU------------GCAACAACAUGUUGCUGGC---GGUUCGGUUUGGAAUUGCUUUUCGAACU-UUUU (((....((..-...))....((((((((((((.(((.(((((....))))).------------))).)))))))))))))))---.....(((((((((.....)))))))))-.... ( -41.60) >DroEre_CAF1 822 117 - 1 GCCCAAAGCAAAAAGGCGAGGAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCUUCGCUUGUAGCAGCAACAACAUGUUGCUGGC---GGUUCGUUUUGGAAUUGCUUUUGGAUCUCUUUU (((...........)))(((((((((((((((((((((((((((((....)))..)).))))))))))...))))))))))(((---(((((......))))))))......)))).... ( -48.50) >DroYak_CAF1 9826 115 - 1 GCCAAAAGCAA-ACGGCAAGGAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCUUCGGUAGUAGCGGCAACAACAUGUUGCUGGU---GAAUCGAUUUGGAAUUGCUUUUGGAUCU-UUUU .((((((((((-....((...(((((((((((((((((((...(((....)))...))))))))(....))))))))))))..)---)..((......)).))))))))))....-.... ( -47.30) >consensus GCCAAAAGCAA_ACGGCGAGGAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCUUCG_U_G_AGC_GCAACAACAUGUUGCUGGC___GGAUCGGUUUGGAAUUGCUUUUCGAACU_UUUU .(.((((((((..........((((((((((((.((((((......................)))))).)))))))))))).........((......)).)))))))).)......... (-27.19 = -28.71 + 1.52)

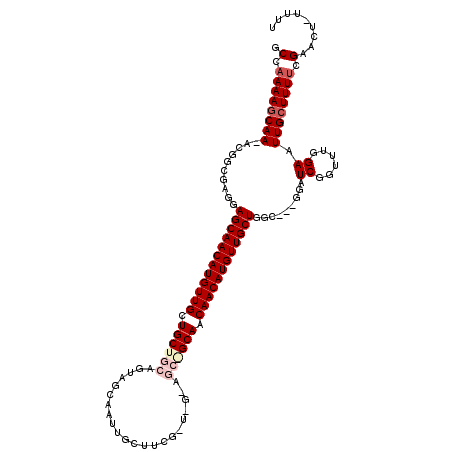
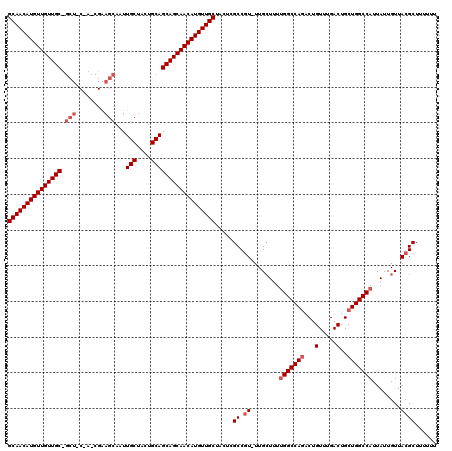
| Location | 5,230,711 – 5,230,830 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -45.06 |
| Consensus MFE | -37.76 |
| Energy contribution | -39.11 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5230711 119 + 22407834 GCAACAUGUUGUUGCUGCUGCUACCGAAGCAUUUGCUACUGCAGCAGCAACAUGUUGCUUCUAGCCGU-UUGAUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUU ((((((((((((((((((((((.....)))).........))))))))))))))))))....(((.((-.((((..(((((((...(.....)..)))))))..))))..)))))..... ( -50.50) >DroSec_CAF1 8065 104 + 1 GCAACAUGUUGUUGC---------------AAUUGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGU-UUGCUUUUGGCCAAACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUU (((((((((((((((---------------...(((....)))))))))))))))))).....((.((-..((...((((((....((......)))))))).....)).))))...... ( -38.00) >DroSim_CAF1 8982 107 + 1 GCAACAUGUUGUUGC------------AGCAAUUGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGU-UUGCUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUU (((((((((((((((------------.(((........))).))))))))))))))).....((.((-..((...(((((((...(.....)..))))))).....)).))))...... ( -43.20) >DroEre_CAF1 859 120 + 1 GCAACAUGUUGUUGCUGCUACAAGCGAAGCAAUUGCUACUGCAGCAGCAACAUGUUGCUCCUCGCCUUUUUGCUUUGGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUU ((((((((((((((((((....(((((.....)))))...)))))))))))))))))).....((............((((((...(.....)..))))))...........))...... ( -48.80) >DroYak_CAF1 9862 119 + 1 GCAACAUGUUGUUGCCGCUACUACCGAAGCAAUUGCUACUGCAGCAGCAACAUGUUGCUCCUUGCCGU-UUGCUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUU (((((((((((((((.(((........)))...(((....)))))))))))))))))).....((.((-..((...(((((((...(.....)..))))))).....)).))))...... ( -44.80) >consensus GCAACAUGUUGUUGC_GCU_C_A_CGAAGCAAUUGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGU_UUGCUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUU (((((((((((((((.(((........)))...(((....)))))))))))))))))).....((.((........(((((((...(.....)..)))))))........))))...... (-37.76 = -39.11 + 1.35)


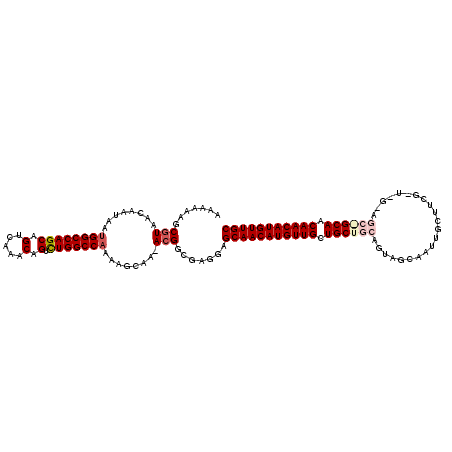
| Location | 5,230,711 – 5,230,830 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -31.80 |
| Energy contribution | -33.16 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5230711 119 - 22407834 AAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAUCAA-ACGGCUAGAAGCAACAUGUUGCUGCUGCAGUAGCAAAUGCUUCGGUAGCAGCAGCAACAACAUGUUGC .....(((..........((((((((.(.....).).))))))).......-...)))....((((((((((((((((((.(.(((....))).)....)))))))))...))))))))) ( -44.35) >DroSec_CAF1 8065 104 - 1 AAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUUUGGCCAAAAGCAA-ACGGCGAGUAGCAACAUGUUGCUGCUGCAGUAGCAAUU---------------GCAACAACAUGUUGC ......((..........((((((((.(.....).).)))))))...((..-...))..)).(((((((((((.((((((....)))...---------------))).))))))))))) ( -32.10) >DroSim_CAF1 8982 107 - 1 AAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAGCAA-ACGGCGAGUAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCU------------GCAACAACAUGUUGC ......((..........((((((((.(.....).).)))))))...((..-...))..)).(((((((((((.(((.(((((....))))).------------))).))))))))))) ( -38.80) >DroEre_CAF1 859 120 - 1 AAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCCAAAGCAAAAAGGCGAGGAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCUUCGCUUGUAGCAGCAACAACAUGUUGC ...................(((((((.(.....).).))))))....((......)).....((((((((((((((((((((((((....)))..)).))))))))))...))))))))) ( -42.70) >DroYak_CAF1 9862 119 - 1 AAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAGCAA-ACGGCAAGGAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCUUCGGUAGUAGCGGCAACAACAUGUUGC ..................((((((((.(.....).).)))))))...((..-...)).....((((((((((((((((((...(((....)))...))))))))(....))))))))))) ( -44.30) >consensus AAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAGCAA_ACGGCGAGGAGCAACAUGUUGCUGCUGCAGUAGCAAUUGCUUCG_U_G_AGC_GCAACAACAUGUUGC .......(((........((((((((.(.....).).)))))))........))).......(((((((((((.((((((......................)))))).))))))))))) (-31.80 = -33.16 + 1.36)

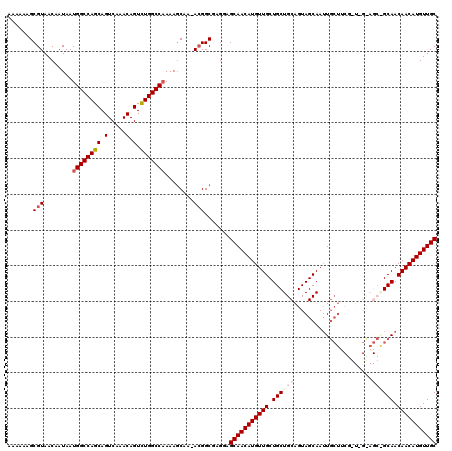
| Location | 5,230,751 – 5,230,861 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -29.34 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5230751 110 + 22407834 GCAGCAGCAACAUGUUGCUUCUAGCCGU-UUGAUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUUGGUCACCUGAUGAUGAAUCAAACAUUGGCGA--------- ((((((......)))))).....(((((-(((((..(((((((...(.....)..)))))))((((..(((.(((.....)))....)))..)))))))))))...)))..--------- ( -34.20) >DroSec_CAF1 8090 119 + 1 GCAGCAGCAACAUGUUGCUACUCGCCGU-UUGCUUUUGGCCAAACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUUGGUCACCUGAUGAUGAAUCAAACAUUGGCGAACGGCCACC ((((((......)))))).....(((((-(((((..((((((((..((.((((..............)))).))...))))))))..((((.....))))......)))))))))).... ( -37.74) >DroSim_CAF1 9010 119 + 1 GCAGCAGCAACAUGUUGCUACUCGCCGU-UUGCUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUUGGUCACCUGAUGAUGAAUCAAACAUUGGCGAACGGCCACC ((((((......)))))).....(((((-(((((..(((((((...(.....)..)))))))((((..(((.(((.....)))....)))..))))..........)))))))))).... ( -40.60) >DroEre_CAF1 899 119 + 1 GCAGCAGCAACAUGUUGCUCCUCGCCUUUUUGCUUUGGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUUGGUCACCUGAUGAUGAAUCAAACAUUGGCGACCGGCCAC- ((((((......)))))).....(((...........((((((...(.....)..))))))...................((((.((.((((.(......).)))))).)))))))...- ( -34.30) >DroYak_CAF1 9902 118 + 1 GCAGCAGCAACAUGUUGCUCCUUGCCGU-UUGCUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUUGGUCACCUGAUGAUGAAUCAAACAUUGGCGACCGGCCAC- ((((((......)))))).....(((((-..((...(((((((...(.....)..))))))).....)).))........((((.((.((((.(......).)))))).)))))))...- ( -36.30) >consensus GCAGCAGCAACAUGUUGCUACUCGCCGU_UUGCUUUUGGCCAGACUGUUUGACUGCUGGCCAUUAUUGUUACGCUUUUUUGGUCACCUGAUGAUGAAUCAAACAUUGGCGAACGGCCAC_ ((((((......))))))...(((((...........((((((...(.....)..)))))).((((..(((.(((.....)))....)))..))))..........)))))......... (-29.34 = -29.62 + 0.28)

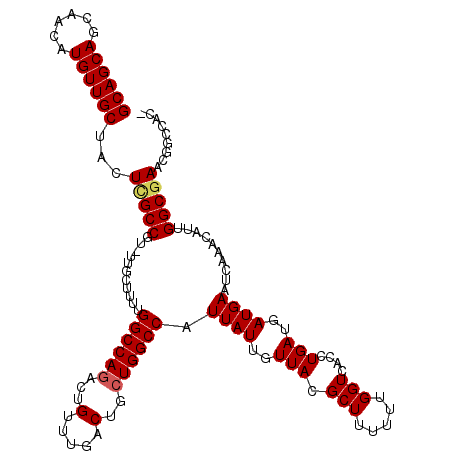

| Location | 5,230,751 – 5,230,861 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -37.46 |
| Consensus MFE | -29.86 |
| Energy contribution | -31.30 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5230751 110 - 22407834 ---------UCGCCAAUGUUUGAUUCAUCAUCAGGUGACCAAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAUCAA-ACGGCUAGAAGCAACAUGUUGCUGCUGC ---------..(((...((((((((((((....)))))....................((((((((.(.....).).)))))))..)))))-)))))(((.(((((....))))).))). ( -31.60) >DroSec_CAF1 8090 119 - 1 GGUGGCCGUUCGCCAAUGUUUGAUUCAUCAUCAGGUGACCAAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUUUGGCCAAAAGCAA-ACGGCGAGUAGCAACAUGUUGCUGCUGC ....((((((.((..((((((...(((((....)))))......))))))........((((((((.(.....).).)))))))...)).)-))))).((((((((....)))))))).. ( -38.90) >DroSim_CAF1 9010 119 - 1 GGUGGCCGUUCGCCAAUGUUUGAUUCAUCAUCAGGUGACCAAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAGCAA-ACGGCGAGUAGCAACAUGUUGCUGCUGC ....((((((.((..((((((...(((((....)))))......))))))........((((((((.(.....).).)))))))...)).)-))))).((((((((....)))))))).. ( -41.30) >DroEre_CAF1 899 119 - 1 -GUGGCCGGUCGCCAAUGUUUGAUUCAUCAUCAGGUGACCAAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCCAAAGCAAAAAGGCGAGGAGCAACAUGUUGCUGCUGC -.((((((((((((..((..(((....))).)))))))))......(.....)......)))))(((((.......(((.(((...........))).)))(((((....)))))))))) ( -36.90) >DroYak_CAF1 9902 118 - 1 -GUGGCCGGUCGCCAAUGUUUGAUUCAUCAUCAGGUGACCAAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAGCAA-ACGGCAAGGAGCAACAUGUUGCUGCUGC -...((((((((((..((..(((....))).)))))))))..................((((((((.(.....).).))))))).......-..))).((.(((((....))))).)).. ( -38.60) >consensus _GUGGCCGGUCGCCAAUGUUUGAUUCAUCAUCAGGUGACCAAAAAAGCGUAACAAUAAUGGCCAGCAGUCAAACAGUCUGGCCAAAAGCAA_ACGGCGAGGAGCAACAUGUUGCUGCUGC .(((((...(((((.((((((...(((((....)))))......))))))........((((((((.(.....).).)))))))..........)))))..(((((....)))))))))) (-29.86 = -31.30 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:03 2006