
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,229,704 – 5,230,055 |
| Length | 351 |
| Max. P | 0.998033 |

| Location | 5,229,704 – 5,229,824 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.35 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -19.47 |
| Energy contribution | -20.04 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229704 120 + 22407834 AAAAAUCUUAUCGGCCAGCGACUUUAAUGUCGGACAUUGAAUUACAGCUCGAGAGCAAAAAAAAAAGAGAUAUAUAAGACACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAU ........(((((((...((((......))))..((((((((((..(((....))).....................(.((.....))).))))))))))((....)).))))))).... ( -25.60) >DroSec_CAF1 7086 119 + 1 ACAAAUCUUAUCGGACAGCGACUUUAAUGUCGGACAUUGAAUUACAGUUCGAGAGCAAAAAA-AAAAAUAUAUAUAAGAAACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAU ........((((((...(((((......)))...(((((((((((.....)...(((((...-.....................))))).))))))))))......))..)))))).... ( -21.16) >DroSim_CAF1 7999 119 + 1 AAAAAUCUUAUCGGCCAGCGACUUUAAUGUCGGACAUUGAAUUACAGUUCGAGACCAAAAAA-AGAGAUAUAUAUAAGAAACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAU ........(((((((...((((......))))..(((((((((((((...............-......................)))..))))))))))((....)).))))))).... ( -21.29) >DroYak_CAF1 8878 120 + 1 AAAAAUCUUAUCGGCCAACGACUUUGAUGGCGGACAUUGAAUUACCGAUCGAGAGCAGAAAAAAAAAAAUUAUAUAAGACAAAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAU ..........((.((((.((....)).)))).))((((((((((.(......).(((((.........................))))).)))))))))).((((....))))....... ( -23.31) >consensus AAAAAUCUUAUCGGCCAGCGACUUUAAUGUCGGACAUUGAAUUACAGUUCGAGAGCAAAAAA_AAAAAUAUAUAUAAGAAACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAU ........(((((((...((((......))))..(((((((((((.....)...(((((.........................))))).))))))))))((....)).))))))).... (-19.47 = -20.04 + 0.56)



| Location | 5,229,744 – 5,229,864 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -17.15 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229744 120 + 22407834 AUUACAGCUCGAGAGCAAAAAAAAAAGAGAUAUAUAAGACACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAUUUUUAUUGCGAUUUUAUCAAGAAAUCGAAAAUGAGCAAAC ......(((((...((((.(((((..(((.(((.((((......)))).))).))).(((.((((....)))).)))..))))).))))((((((.....)))))).....))))).... ( -22.70) >DroSec_CAF1 7126 119 + 1 AUUACAGUUCGAGAGCAAAAAA-AAAAAUAUAUAUAAGAAACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAUUUUUAUUGCGAUUUUAUCAAGAAAUCGAAAAUGAGCAAAC ......(((((...((((.(((-((..((((.................(((......))).((((....))))))))..))))).))))((((((.....)))))).....))))).... ( -19.80) >DroSim_CAF1 8039 119 + 1 AUUACAGUUCGAGACCAAAAAA-AGAGAUAUAUAUAAGAAACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAUUUUUAUUGCGAUUUUAUCAAGAAAUCGAAAAUGAGCAAAA ......(((((...........-((((((.(((((.............(((......))).((((....)))))))))))))))....(((((((.....)))))))....))))).... ( -17.90) >DroYak_CAF1 8918 120 + 1 AUUACCGAUCGAGAGCAGAAAAAAAAAAAUUAUAUAAGACAAAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAUUUUUAUUGCGAUUUUAUCAAGAAAUCGAAAAUGAGCAAAC ..............((........(((((((((((.............(((......))).((((....)))))))))))))))....(((((((.....))))))).......)).... ( -19.30) >consensus AUUACAGUUCGAGAGCAAAAAA_AAAAAUAUAUAUAAGAAACAAUUUGCAUAAUUCAAUGGCAGCAGCAGCUGAUAUAAUUUUUAUUGCGAUUUUAUCAAGAAAUCGAAAAUGAGCAAAC ......(((((.............(((((.(((((.............(((......))).((((....))))))))).)))))....(((((((.....)))))))....))))).... (-17.15 = -17.27 + 0.13)

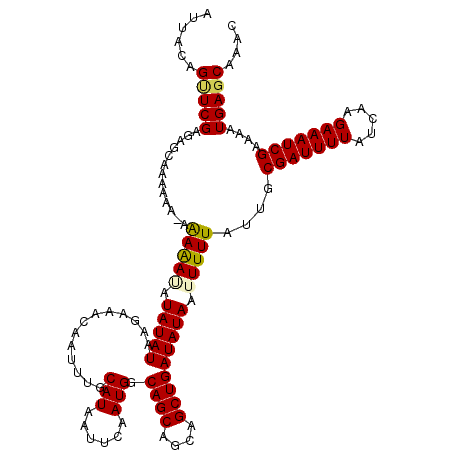
| Location | 5,229,864 – 5,229,983 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
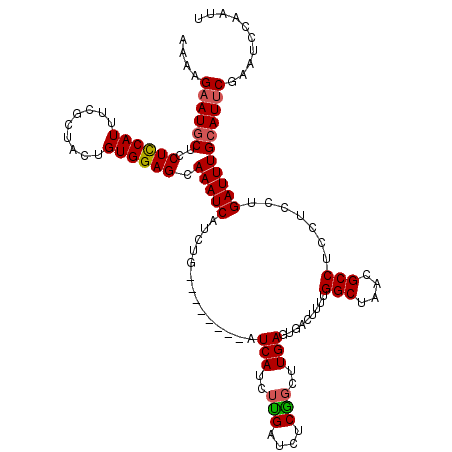
>2L_DroMel_CAF1 5229864 119 + 22407834 CGAAUCGGGGGAUCAAUGACAUUAAUAUUGAUAUUGGGGCGAUGAUUGCCAAUCAUCAUCGCC-GCAAGAUCGAAAUUGGAAUUGGAUUCGAAUGCAAAUCAGGAGGAGGCGUUAGCCAA ((((((.((...(((((..........(((((.(((.(((((((((........)))))))))-.))).))))).)))))..)).)))))).................(((....))).. ( -36.00) >DroSec_CAF1 7245 120 + 1 CGAAUCGGGGGAUCAACGACAUUAAUAUUGAUAUUGGGGCGAUGAUUGCCAAUCAUCAACGCCAGCAAGAUCGAAAUUGGAUUUGGAUUCGAAUGCAAAUCAGGAGGAGGCGUUAGCCAA (((((((..(......)..).(((((.(((((.(((.((((.((((........)))).))))..))).))))).))))).....)))))).................(((....))).. ( -29.70) >DroSim_CAF1 8158 120 + 1 CGAAUCGGGGGAUCAACGACAUUAAUAUUGAUAUUGGGGCGAUGAUUGCCAAUCAUCAUCGCCAGCAAGAUCGAAAUUGGAAUUGGAUUCGAAUGCAAAUCAGGAGGAGGCGUUAGCCAA ((((((((....))..(((..(((((.(((((.(((.(((((((((........)))))))))..))).))))).)))))..))))))))).................(((....))).. ( -34.60) >DroEre_CAF1 11 119 + 1 CAAAUCGGAGGAUCAACGACAUUAAUAUUGAUAUUGGGGCGAUGAUUGCCAAUCAUCAUCGCC-GCAAGAUCGAAAUUGUAAUUGGAUUCGAAUGCAAAUCAGGAGGAGGCGUUAGCCAA ......((.(((((........((((.(((((.(((.(((((((((........)))))))))-.))).))))).))))......))))).(((((...((.....)).)))))..)).. ( -35.34) >DroYak_CAF1 9038 119 + 1 CGAAUCGGGGGAUCAACGACAUUAAUAUUGAUAUUGGGGCGAUGAUUGCCAAUCAUCAUCGCC-GCAAGAUCGAAAUUGUAAUUGGAUUCGAAUGCAAAUCGGUAGGAGGCGUUAGCCAA ((((((((....))..(((...((((.(((((.(((.(((((((((........)))))))))-.))).))))).))))...)))))))))..(((......)))...(((....))).. ( -36.50) >consensus CGAAUCGGGGGAUCAACGACAUUAAUAUUGAUAUUGGGGCGAUGAUUGCCAAUCAUCAUCGCC_GCAAGAUCGAAAUUGGAAUUGGAUUCGAAUGCAAAUCAGGAGGAGGCGUUAGCCAA ......((.(((((........((((.(((((.....(((((((((........)))))))))......))))).))))......))))).(((((...((.....)).)))))..)).. (-28.80 = -29.00 + 0.20)


| Location | 5,229,864 – 5,229,983 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229864 119 - 22407834 UUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUUCCAAUUUCGAUCUUGC-GGCGAUGAUGAUUGGCAAUCAUCGCCCCAAUAUCAAUAUUAAUGUCAUUGAUCCCCCGAUUCG ..(((....))).................((((((.............(((.(((.-(((((((((........))))))))).))).)))...((((((...)))))).....)))))) ( -30.80) >DroSec_CAF1 7245 120 - 1 UUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAAUCCAAUUUCGAUCUUGCUGGCGUUGAUGAUUGGCAAUCAUCGCCCCAAUAUCAAUAUUAAUGUCGUUGAUCCCCCGAUUCG ..(((.((((((............(((.(((((............)))))...))).))))))((((((.....))))))))).....((((((.........))))))........... ( -25.02) >DroSim_CAF1 8158 120 - 1 UUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUUCCAAUUUCGAUCUUGCUGGCGAUGAUGAUUGGCAAUCAUCGCCCCAAUAUCAAUAUUAAUGUCGUUGAUCCCCCGAUUCG ..(((....))).................((((((.............(((.(((..(((((((((........))))))))).))).)))...((((((...)))))).....)))))) ( -29.40) >DroEre_CAF1 11 119 - 1 UUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUUACAAUUUCGAUCUUGC-GGCGAUGAUGAUUGGCAAUCAUCGCCCCAAUAUCAAUAUUAAUGUCGUUGAUCCUCCGAUUUG ..(((....))).......((((((....))))))((((.(((.....(((.(((.-(((((((((........))))))))).))).)))........))).))))............. ( -30.22) >DroYak_CAF1 9038 119 - 1 UUGGCUAACGCCUCCUACCGAUUUGCAUUCGAAUCCAAUUACAAUUUCGAUCUUGC-GGCGAUGAUGAUUGGCAAUCAUCGCCCCAAUAUCAAUAUUAAUGUCGUUGAUCCCCCGAUUCG ..(((....))).................((((((.............(((.(((.-(((((((((........))))))))).))).)))...((((((...)))))).....)))))) ( -31.10) >consensus UUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUUCCAAUUUCGAUCUUGC_GGCGAUGAUGAUUGGCAAUCAUCGCCCCAAUAUCAAUAUUAAUGUCGUUGAUCCCCCGAUUCG ..(((....)))............(((.(((((............)))))...))).(((((((((........))))))))).....((((((.........))))))........... (-27.08 = -27.12 + 0.04)


| Location | 5,229,943 – 5,230,055 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -23.16 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229943 112 - 22407834 AAAAGAAUGCUCCUCCAUUUCGCUACUGUGGAGCAAAUCGUCUG--------AUCAUCUUGAUCUCGGCUUGAGUGACUUUUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUU ....((((((..((((((.........)))))).((((((((.(--------(((.....))))..)))..(((.((.....(((....))))))))..))))))))))).......... ( -30.80) >DroSec_CAF1 7325 112 - 1 AACAGAAUGCUCCUCCAUUUCGCUACUGUGGAGCAAAUCAUCUG--------GUCAUCACGAUCUCGGCUUGAGUGACUUUUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAAU ....((((((..((((((.........)))))).((((((...(--------(((((((((....))...))).)))))...(((....)))......)))))))))))).......... ( -29.60) >DroSim_CAF1 8238 112 - 1 AACAGAAUACUCCUCCAUUUCGCUACUGUGGAGCAAAUCAUCUG--------GUCAUCUUGAUCUCGGCUUGAGUGACUUUUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUU ....((((.....(((((.........)))))((((((((...(--------((((.((..(.......)..)))))))...(((....)))......)))))))))))).......... ( -28.70) >DroEre_CAF1 90 120 - 1 AAAAGAAUGCUCCUCCAUUUCGCUACUGUGGAGCAAAUCAUCUGAUCAUCUGAUCAUCUUGAUUUCAGCUUGAGUGACUUUUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUU ....((((((..((((((.........))))))..(((((..(((((....)))))...)))))((((...(((.((.....(((....))))))))))))...)))))).......... ( -32.10) >DroYak_CAF1 9117 112 - 1 AUAAGUAUGCUCCUUCAUUUCGCUACUGUGGAGCAAAUCAUCUG--------AUCAUUUUGAUCUCAGCUUGAGUGACUUUUGGCUAACGCCUCCUACCGAUUUGCAUUCGAAUCCAAUU .............(((..(((((....)))))(((((((.((..--------.(((..(((....)))..)))..)).....(((....))).......)))))))....)))....... ( -24.10) >consensus AAAAGAAUGCUCCUCCAUUUCGCUACUGUGGAGCAAAUCAUCUG________AUCAUCUUGAUCUCGGCUUGAGUGACUUUUGGCUAACGCCUCCUCCUGAUUUGCAUUCGAAUCCAAUU ....((((((..((((((.........)))))).(((((..............(((..(((....)))..))).........(((....))).......))))))))))).......... (-23.16 = -23.20 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:56 2006