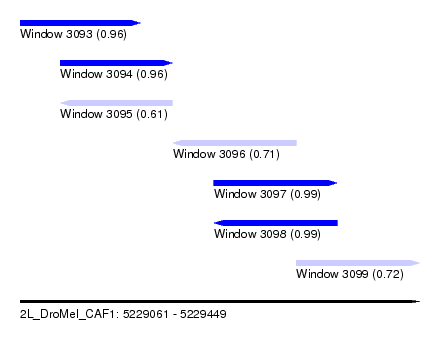
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,229,061 – 5,229,449 |
| Length | 388 |
| Max. P | 0.988805 |

| Location | 5,229,061 – 5,229,178 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.34 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -27.11 |
| Energy contribution | -28.68 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229061 117 + 22407834 CCAAA-CAAAUCUCACUUGCCAAUGAGCAGAUGGUCGAUGCUAAUGGUCAGAUUU--GUGUCUGACACAAUUUGGCAUAUGGACUAUCCCCAAUCUGCCCUCGCCUUCGUUUGGGCAAUU .....-.....((((........))))..(((((((.((((((((((((((((..--..))))))).))..)))))))...))))))).......(((((..((....))..)))))... ( -36.80) >DroSec_CAF1 6470 106 + 1 CCGAA-CAAAUCUCACUU-CCAAUGAGCAGAUGGUCGAUGCUAAUGGCCAGAUUUCUGUGCCUGACACAAUUUGGCAUAUGGACUAGCCCCAAUCUGCCCUCG------------CAAUU .(((.-......(((...-....)))((((((((((.(((((((.((((((....))).))).........)))))))...)))).......))))))..)))------------..... ( -24.01) >DroSim_CAF1 7366 118 + 1 CCGAA-CAAAUCUCACUU-CCAAUGAGCAGAUGGUCGAUGCUAAUGGUCAGAUUUUUGUAUCUGACACAAUUUGGCAUAUGGACUAGCCCCAAUCUGCCCUCGCCUUCACUUGGGCAAUU .....-............-.......((((((((((.((((((((((((((((......))))))).))..)))))))...)))).......))))))....((((......)))).... ( -33.21) >DroYak_CAF1 7599 117 + 1 CCAAAGCAAACCUCACUU-ACAAUGAGCAGAUGGUCGAUGCUAAUGGUCAGAUUU--GUGUCUGACACAAUUUGGCAUAUGGACUAUCCCCAAUCUGCCCUCUCCUUCACUCGGGCAAUU ...........((((...-....))))..(((((((.((((((((((((((((..--..))))))).))..)))))))...))))))).......(((((............)))))... ( -34.80) >consensus CCAAA_CAAAUCUCACUU_CCAAUGAGCAGAUGGUCGAUGCUAAUGGUCAGAUUU__GUGUCUGACACAAUUUGGCAUAUGGACUAGCCCCAAUCUGCCCUCGCCUUCACUUGGGCAAUU ...........((((........))))..(((((((.((((((((((((((((......))))))).))..)))))))...))))))).......(((((............)))))... (-27.11 = -28.68 + 1.56)

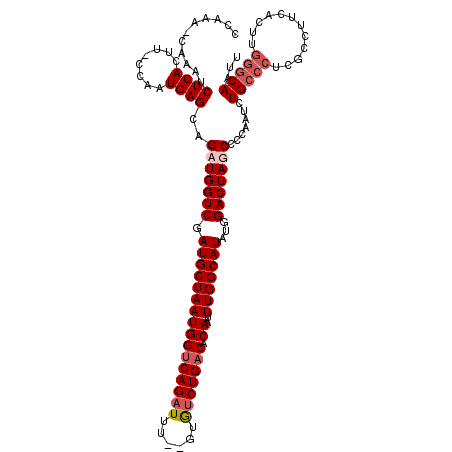
| Location | 5,229,100 – 5,229,209 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -22.45 |
| Energy contribution | -24.08 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229100 109 + 22407834 CUAAUGGUCAGAUUU--GUGUCUGACACAAUUUGGCAUAUGGACUAUCCCCAAUCUGCCCUCGCCUUCGUUUGGGCAAUUAAAAACAAUUGC---------ACAUCUGCGAUAGUGCAUA ......((((((((.--(((.....))))))))))).((((.((((((.......(((((..((....))..))))).............((---------(....))))))))).)))) ( -32.40) >DroSec_CAF1 6508 108 + 1 CUAAUGGCCAGAUUUCUGUGCCUGACACAAUUUGGCAUAUGGACUAGCCCCAAUCUGCCCUCG------------CAAUUAAAAACAAUUGCAGCAUUCAUACAUCUGCGAUGGUGCAUA .....(((.((.((..((((((...........))))))..)))).)))......((((((((------------(((((......)))))).(((..........))))).)).))).. ( -27.20) >DroSim_CAF1 7404 120 + 1 CUAAUGGUCAGAUUUUUGUAUCUGACACAAUUUGGCAUAUGGACUAGCCCCAAUCUGCCCUCGCCUUCACUUGGGCAAUUAAAAACAAUUGCAGCAUUCAUACAUCUGCGAUAGUGCAUA ....(((((((((......))))))).))....((((..(((.......)))...))))...((((......))))........((.(((((((.((......))))))))).))..... ( -30.40) >DroYak_CAF1 7638 118 + 1 CUAAUGGUCAGAUUU--GUGUCUGACACAAUUUGGCAUAUGGACUAUCCCCAAUCUGCCCUCUCCUUCACUCGGGCAAUUAAAAACAAUUGCAGUAUUCAUGCAUCUGCGAUAGUGCAUA ......((((((((.--(((.....))))))))))).((((.((((((.......(((((............)))))........((..((((.......))))..)).)))))).)))) ( -33.00) >consensus CUAAUGGUCAGAUUU__GUGUCUGACACAAUUUGGCAUAUGGACUAGCCCCAAUCUGCCCUCGCCUUCACUUGGGCAAUUAAAAACAAUUGCAGCAUUCAUACAUCUGCGAUAGUGCAUA ......((((((((...(((.....))))))))))).((((.((((((.......(((((............))))).............((((...........)))))))))).)))) (-22.45 = -24.08 + 1.62)

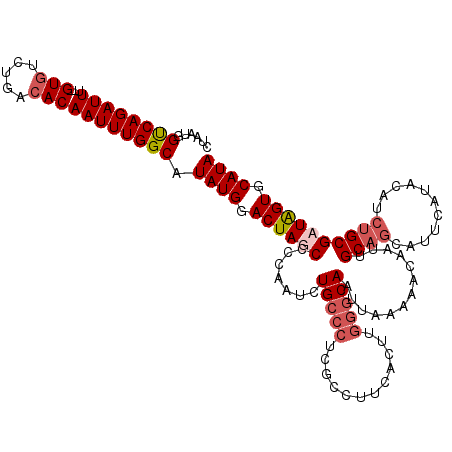
| Location | 5,229,100 – 5,229,209 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.85 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229100 109 - 22407834 UAUGCACUAUCGCAGAUGU---------GCAAUUGUUUUUAAUUGCCCAAACGAAGGCGAGGGCAGAUUGGGGAUAGUCCAUAUGCCAAAUUGUGUCAGACAC--AAAUCUGACCAUUAG ((((.((((((.(.(((.(---------((..((((((((..((....))..))))))))..))).))).).)))))).))))........((.((((((...--...)))))))).... ( -31.70) >DroSec_CAF1 6508 108 - 1 UAUGCACCAUCGCAGAUGUAUGAAUGCUGCAAUUGUUUUUAAUUG------------CGAGGGCAGAUUGGGGCUAGUCCAUAUGCCAAAUUGUGUCAGGCACAGAAAUCUGGCCAUUAG ..(((......)))((..((......(((((((((....))))))------------).))((((...((((.....))))..))))....))..)).(((.(((....))))))..... ( -31.10) >DroSim_CAF1 7404 120 - 1 UAUGCACUAUCGCAGAUGUAUGAAUGCUGCAAUUGUUUUUAAUUGCCCAAGUGAAGGCGAGGGCAGAUUGGGGCUAGUCCAUAUGCCAAAUUGUGUCAGAUACAAAAAUCUGACCAUUAG ..(((.((.((((....(((....))).(((((((....)))))))....)))))))))..((((...((((.....))))..))))....((.(((((((......))))))))).... ( -32.30) >DroYak_CAF1 7638 118 - 1 UAUGCACUAUCGCAGAUGCAUGAAUACUGCAAUUGUUUUUAAUUGCCCGAGUGAAGGAGAGGGCAGAUUGGGGAUAGUCCAUAUGCCAAAUUGUGUCAGACAC--AAAUCUGACCAUUAG ((((.((((((((((.((((.......)))).)))).(((((((((((............))))).)))))))))))).))))........((.((((((...--...)))))))).... ( -32.10) >consensus UAUGCACUAUCGCAGAUGUAUGAAUGCUGCAAUUGUUUUUAAUUGCCCAAGUGAAGGCGAGGGCAGAUUGGGGAUAGUCCAUAUGCCAAAUUGUGUCAGACAC__AAAUCUGACCAUUAG ..(((......)))((((..........((((((........(((((........))))).((((...((((.....))))..)))).))))))((((((........)))))))))).. (-25.85 = -25.85 + -0.00)

| Location | 5,229,209 – 5,229,329 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -32.00 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229209 120 - 22407834 AUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUCGGCUGCCGUAUGCAGAGGAAAAAUGUCGGAUAAAAAUAUAGGAAAAU ........((....((((.(((((((((((((.((((...))))...((......))..))))))))))))).)))).((((((...........))).)))............)).... ( -29.90) >DroSec_CAF1 6616 120 - 1 AUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUUGGCUGCCGUAUGCAGAGAGAAAAUGUCGGAUAAAAAUAUAGGAAAAU ........((....((((((((((((((((((.((((...))))...((......))..)))))))))))))))))).((((((...........))).)))............)).... ( -34.00) >DroSim_CAF1 7524 120 - 1 AUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUUGGCUGCCGUAUGCAGAGGGAAAAUGUCGGAUAAAAAUAUAGGAAAAU ........((....((((((((((((((((((.((((...))))...((......))..)))))))))))))))))).((((((...........))).)))............)).... ( -34.00) >DroYak_CAF1 7756 120 - 1 AUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUCGGCUGCCGUAUGCAGAGGGAAAAUGUCGGAUAAAAAUAUAUGAAAAU ........((((..((((.(((((((((((((.((((...))))...((......))..))))))))))))).)))).((((((...........))).)))..........)))).... ( -31.20) >consensus AUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUCGGCUGCCGUAUGCAGAGGGAAAAUGUCGGAUAAAAAUAUAGGAAAAU ........((....((((((((((((((((((.((((...))))...((......))..)))))))))))))))))).((((((...........))).)))............)).... (-32.00 = -32.50 + 0.50)

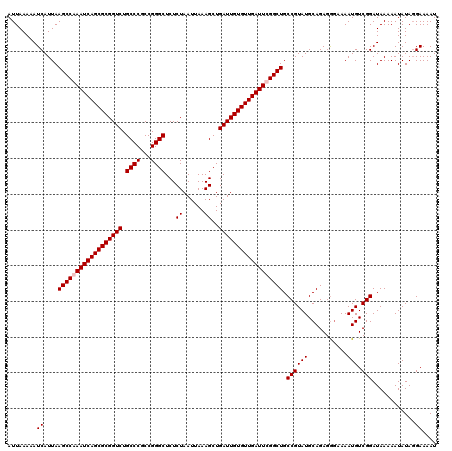
| Location | 5,229,249 – 5,229,369 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.04 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229249 120 + 22407834 GGCAGCCGAAUCAACACAAUCAGCUUUAAUUAGAGAGCCCGGCGGGCAGACCGCGCUGAUUUGGCUUAAUGAUUUUUAAUUGCAGGCGCAAAUUAUUAAAAAACAUUCGACAAAAAAAAA ...((((((((((.(.......(((((......)))))...((((.....))))).)))))))))).((((.(((((((((((....)))....)))))))).))))............. ( -31.90) >DroSec_CAF1 6656 117 + 1 GGCAGCCAAAUCAACACAAUCAGCUUUAAUUAGAGAGCCCGGCGGGCAGACCGCGCUGAUUUGGCUUAAUGAUUUUUAAUUGCAGGCGCAAAUUAUUAAAAAACAUUCGA---AAAAACA ...((((((((((.(.......(((((......)))))...((((.....))))).)))))))))).((((.(((((((((((....)))....)))))))).))))...---....... ( -32.20) >DroSim_CAF1 7564 116 + 1 GGCAGCCAAAUCAACACAAUCAGCUUUAAUUAGAGAGCCCGGCGGGCAGACCGCGCUGAUUUGGCUUAAUGAUUUUUAAUUGCAGGCGCAAAUUAUUAAAAAACAUUCGA---AAAAA-A ...((((((((((.(.......(((((......)))))...((((.....))))).)))))))))).((((.(((((((((((....)))....)))))))).))))...---.....-. ( -32.20) >DroYak_CAF1 7796 116 + 1 GGCAGCCGAAUCAACACAAUCAGCUUUAAUUAGAGAGCCCGGCGGGCAGACCGCGCUGAUUUGGCUUAAUGAUUUUUAAUUGCAGGCGCAAAUUAUUAAAAAACAUUCGC---AAAAA-A .((((((((((((.(.......(((((......)))))...((((.....))))).)))))))))).((((.(((((((((((....)))....)))))))).)))).))---.....-. ( -34.40) >consensus GGCAGCCAAAUCAACACAAUCAGCUUUAAUUAGAGAGCCCGGCGGGCAGACCGCGCUGAUUUGGCUUAAUGAUUUUUAAUUGCAGGCGCAAAUUAUUAAAAAACAUUCGA___AAAAA_A ...((((((((((.(.......(((((......)))))...((((.....))))).)))))))))).((((.(((((((((((....)))....)))))))).))))............. (-32.30 = -32.05 + -0.25)

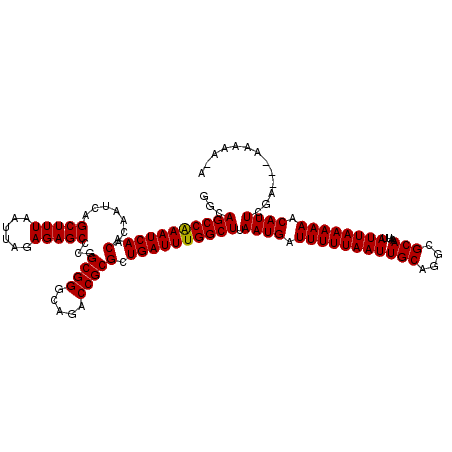
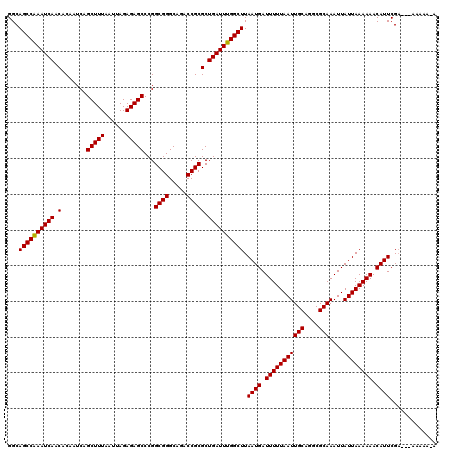
| Location | 5,229,249 – 5,229,369 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.04 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -36.30 |
| Energy contribution | -36.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229249 120 - 22407834 UUUUUUUUUGUCGAAUGUUUUUUAAUAAUUUGCGCCUGCAAUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUCGGCUGCC .............((((.((((((((....(((....))))))))))).)))).((((.(((((((((((((.((((...))))...((......))..))))))))))))).))))... ( -34.20) >DroSec_CAF1 6656 117 - 1 UGUUUUU---UCGAAUGUUUUUUAAUAAUUUGCGCCUGCAAUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUUGGCUGCC .......---...((((.((((((((....(((....))))))))))).)))).((((((((((((((((((.((((...))))...((......))..))))))))))))))))))... ( -38.30) >DroSim_CAF1 7564 116 - 1 U-UUUUU---UCGAAUGUUUUUUAAUAAUUUGCGCCUGCAAUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUUGGCUGCC .-.....---...((((.((((((((....(((....))))))))))).)))).((((((((((((((((((.((((...))))...((......))..))))))))))))))))))... ( -38.30) >DroYak_CAF1 7796 116 - 1 U-UUUUU---GCGAAUGUUUUUUAAUAAUUUGCGCCUGCAAUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUCGGCUGCC .-.....---((.((((.((((((((....(((....))))))))))).)))).((((.(((((((((((((.((((...))))...((......))..))))))))))))).)))))). ( -35.50) >consensus U_UUUUU___UCGAAUGUUUUUUAAUAAUUUGCGCCUGCAAUUAAAAAUCAUUAAGCCAAAUCAGCGCGGUCUGCCCGCCGGGCUCUCUAAUUAAAGCUGAUUGUGUUGAUUCGGCUGCC .............((((.((((((((....(((....))))))))))).)))).((((((((((((((((((.((((...))))...((......))..))))))))))))))))))... (-36.30 = -36.80 + 0.50)



| Location | 5,229,329 – 5,229,449 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5229329 120 + 22407834 UGCAGGCGCAAAUUAUUAAAAAACAUUCGACAAAAAAAAAACACAUGUGCAUAUACAAAUAUGAAAAAUUGCAGCAGCUGUGGACCACCCAGGCGAAGGCGUUUAAAAACGCGCCGAAAU ....(((((..................................(((((..........))))).......((....(((.(((.....))))))....))..........)))))..... ( -22.50) >DroSec_CAF1 6736 114 + 1 UGCAGGCGCAAAUUAUUAAAAAACAUUCGA---AAAAACAGCACAUGUGCAUAUACAAAUAUGAAAAAUUGCAGCAGCUGUGGACCACCCAGGCGAAGGCGUU---AAACGCGCCGAAAA ....(((((.....................---.......(((......(((((....)))))......)))(((..(((.((....)))))((....)))))---....)))))..... ( -26.00) >DroSim_CAF1 7644 116 + 1 UGCAGGCGCAAAUUAUUAAAAAACAUUCGA---AAAAA-AGCACAUGUGCAUAUACAAAUAUGAAAAAUUGCAGCAGCUGUGGACCACCCAGGCGAAGGCGUUUAAAAACGCGCCGAAAA ....(((((.....................---.....-.(((......(((((....)))))......)))(((..(((.((....)))))((....))))).......)))))..... ( -25.40) >DroYak_CAF1 7876 116 + 1 UGCAGGCGCAAAUUAUUAAAAAACAUUCGC---AAAAA-AGUACAUGUGCAUAUACAAAUAUGAAAAAUUGCAGCAGCUGUGGACCACCCCGCCGAAGGCGUUUAAAAACGCGCCGAAAU ....(((((...................((---((...-.(((..((((....))))..)))......)))).((..(.((((......)))).)...))..........)))))..... ( -24.50) >consensus UGCAGGCGCAAAUUAUUAAAAAACAUUCGA___AAAAA_AGCACAUGUGCAUAUACAAAUAUGAAAAAUUGCAGCAGCUGUGGACCACCCAGGCGAAGGCGUUUAAAAACGCGCCGAAAA ....(((((...............................(((......(((((....)))))......))).((.(((.(((.....))))))....))..........)))))..... (-20.64 = -21.20 + 0.56)

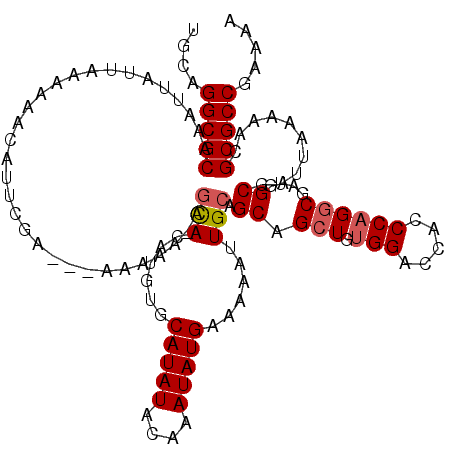
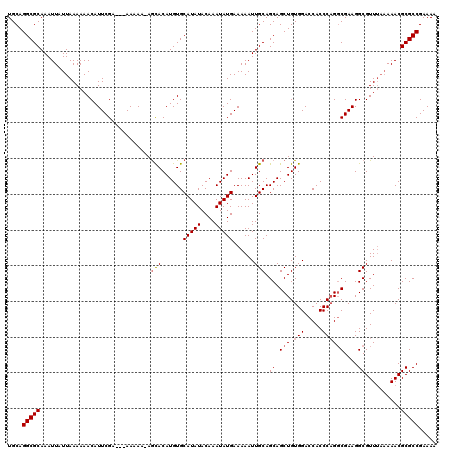
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:51 2006