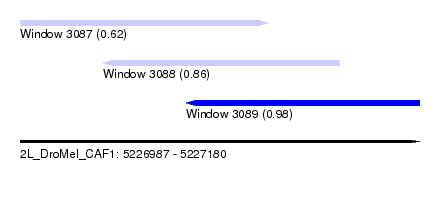
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,226,987 – 5,227,180 |
| Length | 193 |
| Max. P | 0.983953 |

| Location | 5,226,987 – 5,227,107 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -26.51 |
| Energy contribution | -26.32 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5226987 120 + 22407834 AAAUUCUUAAUAAAACGCGCUGCAGUCUCUCGAAAUAUUUGCAAGUGUCUCAACGUCGCAUGCGUUUUAAUUAAGUGUCGCAUAUUUUUAAUUAAAUCACAAACGCCCAGGCGACGCGAC .....((((((((((((((.(((((.............))))).(((.(.....).))).)))))))).)))))).(((((..((((......))))......(((....)))..))))) ( -28.12) >DroSec_CAF1 4437 120 + 1 AAAUUCUUAAUAAAACGCGCUGCAGUCUCUCGAAAUAUUUCCAAGUGUCUUAACGUCGCAUGCGUUUUAAUUAAGUGUCGCAUAUUUCUAAUUAAAUCACAAACGCCCAGGCGACGCGAC .....((((((((((((((.(((........(((....)))...((......))...))))))))))).)))))).(((((.((........)).........(((....)))..))))) ( -26.20) >DroSim_CAF1 5315 120 + 1 AAAUUCUUAAUAAAACGCGCUGCAGUCUCUCGAAAUAUUUGCAAGUGUCUCAACGUCGCAUGCGUUUUAAUUAAGUGUCGCAUAUUUUUAAUUAAAUCACAAACGCCCAGGCGACGCGAC .....((((((((((((((.(((((.............))))).(((.(.....).))).)))))))).)))))).(((((..((((......))))......(((....)))..))))) ( -28.12) >DroYak_CAF1 5511 120 + 1 AAAUUCUUAAUAAAACGCGCUGCAGUCUCUCGAAAUAUUUGCAAGUGUCUCUACGUCGCAUGCGUUUUAAUUAAGUGUCGCAUAUUUUUAAUUAAAUCACAAACGCCCAGGCGACGCGAC .....((((((((((((((.(((((.............))))).(((.(.....).))).)))))))).)))))).(((((..((((......))))......(((....)))..))))) ( -28.12) >consensus AAAUUCUUAAUAAAACGCGCUGCAGUCUCUCGAAAUAUUUGCAAGUGUCUCAACGUCGCAUGCGUUUUAAUUAAGUGUCGCAUAUUUUUAAUUAAAUCACAAACGCCCAGGCGACGCGAC .....((((((((((((((.(((.(.....)((.(((((....))))).))......))))))))))).)))))).(((((..((((......))))......(((....)))..))))) (-26.51 = -26.32 + -0.19)

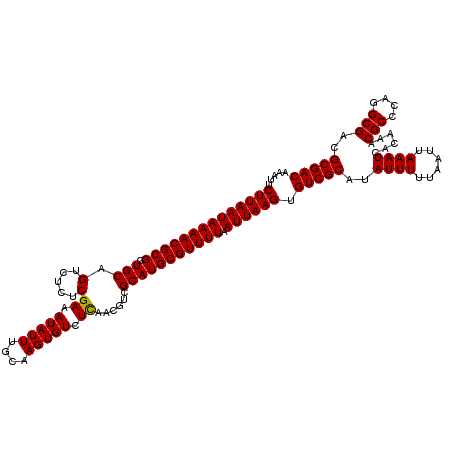
| Location | 5,227,027 – 5,227,141 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -33.41 |
| Energy contribution | -33.47 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5227027 114 - 22407834 CCUCCAG------AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUGCGACACUUAAUUAAAACGCAUGCGACGUUGAGACACUUGC ..(((((------.((..((((((((((((.....)))))))))))).)))))))(((.((((.(((((((((.............))))))))).)))).)))................ ( -35.32) >DroSec_CAF1 4477 114 - 1 CCUCCAG------AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAGAAAUAUGCGACACUUAAUUAAAACGCAUGCGACGUUAAGACACUUGG ..(((((------.((..((((((((((((.....)))))))))))).)))))))(((.((((.(((((((((.............))))))))).)))).)))................ ( -35.02) >DroSim_CAF1 5355 114 - 1 CCUCCAG------AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUGCGACACUUAAUUAAAACGCAUGCGACGUUGAGACACUUGC ..(((((------.((..((((((((((((.....)))))))))))).)))))))(((.((((.(((((((((.............))))))))).)))).)))................ ( -35.32) >DroYak_CAF1 5551 120 - 1 CCUCCAGAGCCAGAGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUGCGACACUUAAUUAAAACGCAUGCGACGUAGAGACACUUGC .(((.....((((.((..((((((((((((.....)))))))))))).))))))(((((((((.(((((((((.............))))))))).))))...))))).)))........ ( -37.82) >consensus CCUCCAG______AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUGCGACACUUAAUUAAAACGCAUGCGACGUUGAGACACUUGC .(((..........((((((((((((((((.....)))))))))))).....))))((.((((.(((((((((.............))))))))).)))).))......)))........ (-33.41 = -33.47 + 0.06)

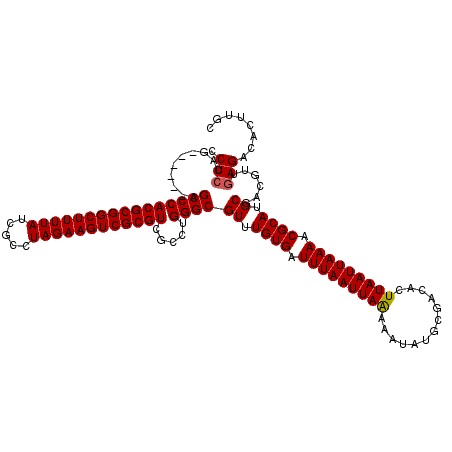

| Location | 5,227,067 – 5,227,180 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.53 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5227067 113 - 22407834 AUUUGCUCGAUU-CAAAUCGCACUCCUCCCUUUUGGAUUCCCUCCAG------AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUG ........((((-.((((((((..(......((((((.....)))))------)((((((((((((((((.....)))))))))))).....)))))..))))))))))))......... ( -35.90) >DroSec_CAF1 4517 110 - 1 AUUU----GAUUCCAAAUCGCACUCCUCCCUUUUGGAUUCCCUCCAG------AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAGAAAUAUG .(((----((((..((((((((..(......((((((.....)))))------)((((((((((((((((.....)))))))))))).....)))))..)))))))))))))))...... ( -37.60) >DroSim_CAF1 5395 114 - 1 AUUUGCUCGAUUCCAAAUCGCACUCCUCCCUUUUGGAUUCCCUCCAG------AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUG ..............((((((((..(......((((((.....)))))------)((((((((((((((((.....)))))))))))).....)))))..))))))))............. ( -35.10) >DroYak_CAF1 5591 119 - 1 AUUUGUGCGAUU-CAAAUCGCACUCCUCCCUUUUGGUUACCCUCCAGAGCCAGAGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUG ....(((((((.-...))))))).......(((((((((...(((((((((((.((..((((((((((((.....)))))))))))).)))).))).)))).))..)))))))))..... ( -40.20) >consensus AUUUGCUCGAUU_CAAAUCGCACUCCUCCCUUUUGGAUUCCCUCCAG______AGCCCACGCGGCUUUUAUCGCCUAGAAGUCGCGUCGCCUGGGCGUUUGUGAUUUAAUUAAAAAUAUG ..............((((((((..(........((((.....))))........((((((((((((((((.....)))))))))))).....)))))..))))))))............. (-32.28 = -32.53 + 0.25)

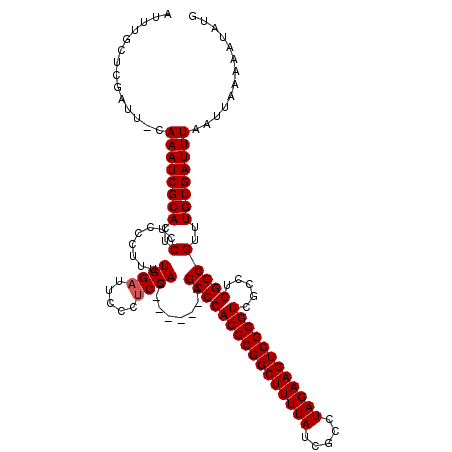
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:42 2006