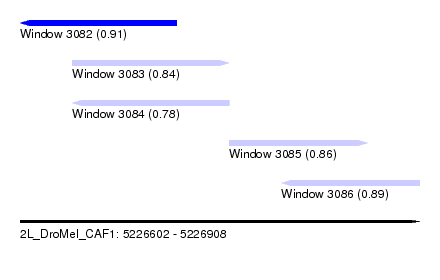
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,226,602 – 5,226,908 |
| Length | 306 |
| Max. P | 0.909389 |

| Location | 5,226,602 – 5,226,722 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -31.39 |
| Energy contribution | -31.32 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5226602 120 - 22407834 CGAAACGCACUCUGUUCGCAAAACAAUGGUGAUUUUACAGUCGCUCGUUGUUGCGCUACAGUGACGCAACAGUGCGUUAACAGUGGGCGUCGAAUGCUACCGAGAGUUCACUGUACCAGC ...((((((((((((.((((..(((((((((((......))))).))))))))))..)))).........)))))))).(((((((((.(((........)))..)))))))))...... ( -44.10) >DroSec_CAF1 4064 107 - 1 CGAAACGCACUCA-CUCGCAAAACAAUGGCGAUUUUACAG------------GCGCUACAGUGACGCAACAGUGCGUUAACAGUGGGCGUUGGAUGCUACCGAGAGUUCACUGUAGCAGG ...((((((((..-.((((.........))))........------------((((......).)))...)))))))).(((((((((.((((......))))..)))))))))...... ( -34.00) >DroSim_CAF1 4941 108 - 1 CGAAACGCACUCUGGUCGCAAAACAAUGGCGAUUUUACAG------------GCGCUACAGUGACGCAACAGUGCGUUAACAGUGGGCGUCGGAUGCUACCGAGAGUUCACUGUAGCAGC ...((((((((((((((((.........)))))....)))------------((((......).)))...)))))))).(((((((((.((((......))))..)))))))))...... ( -38.80) >DroYak_CAF1 5113 120 - 1 CAAAACGCACUCUGGUCGCACAACAAUGGCGAUUUCACAGUCGCUCGAUGUUGAGCUACAGUGACGCAACAGUGCGUUAACAGUGGCCGUCGGAUGCUACUGAGAGUUCACUGUACCAGC ...........((((..((.(((((.(((((((......))))).)).))))).))((((((((((((....))))....(((((((........))))))).....)))))))))))). ( -43.80) >consensus CGAAACGCACUCUGGUCGCAAAACAAUGGCGAUUUUACAG____________GCGCUACAGUGACGCAACAGUGCGUUAACAGUGGGCGUCGGAUGCUACCGAGAGUUCACUGUACCAGC ...((((((((....((((.........))))....................((((......).)))...)))))))).((((((..(.((((......))))..)..))))))...... (-31.39 = -31.32 + -0.06)

| Location | 5,226,642 – 5,226,762 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.15 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5226642 120 + 22407834 UUAACGCACUGUUGCGUCACUGUAGCGCAACAACGAGCGACUGUAAAAUCACCAUUGUUUUGCGAACAGAGUGCGUUUCGCUUUGCAUAAAUGCAUUUUACAAGCGUUUAAUUGACUUUC ..(((((((((((((((.......)))))))..........((((((((.......)))))))).....)))))))).(((((((((....))))......))))).............. ( -30.50) >DroSec_CAF1 4104 107 + 1 UUAACGCACUGUUGCGUCACUGUAGCGC------------CUGUAAAAUCGCCAUUGUUUUGCGAG-UGAGUGCGUUUCGCUUUGCAUAAAUGCAUUUUACAAGCGUUUAAUUGACUUUC .....(((....)))((((....(((((------------.((((((((.((.(((....((((((-((((.....))))).)))))..))))))))))))).)))))....)))).... ( -32.80) >DroSim_CAF1 4981 108 + 1 UUAACGCACUGUUGCGUCACUGUAGCGC------------CUGUAAAAUCGCCAUUGUUUUGCGACCAGAGUGCGUUUCGCUUUGCAUAAAUGCAUUUUACAAGCGUUUAAUUGACUUUC .....(((....)))((((....(((((------------.((((((((.((.(((....(((((...(((.....)))...)))))..))))))))))))).)))))....)))).... ( -29.00) >DroYak_CAF1 5153 120 + 1 UUAACGCACUGUUGCGUCACUGUAGCUCAACAUCGAGCGACUGUGAAAUCGCCAUUGUUGUGCGACCAGAGUGCGUUUUGCUUUGCAUAAAUGCAUUUUACAAGCGUUUAAUUGACUUUC ..(((((.(((.....((((.((.((((......)))).)).))))..((((((....)).)))).)))((((((((((((...))).)))))))))......)))))............ ( -34.60) >consensus UUAACGCACUGUUGCGUCACUGUAGCGC____________CUGUAAAAUCGCCAUUGUUUUGCGACCAGAGUGCGUUUCGCUUUGCAUAAAUGCAUUUUACAAGCGUUUAAUUGACUUUC ..((((((((((((((....))))))...............((((((((.......)))))))).....)))))))).(((((((((....))))......))))).............. (-24.27 = -24.15 + -0.12)


| Location | 5,226,642 – 5,226,762 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5226642 120 - 22407834 GAAAGUCAAUUAAACGCUUGUAAAAUGCAUUUAUGCAAAGCGAAACGCACUCUGUUCGCAAAACAAUGGUGAUUUUACAGUCGCUCGUUGUUGCGCUACAGUGACGCAACAGUGCGUUAA ..............(((((......((((....))))))))).((((((((((((.((((..(((((((((((......))))).))))))))))..)))).........)))))))).. ( -36.60) >DroSec_CAF1 4104 107 - 1 GAAAGUCAAUUAAACGCUUGUAAAAUGCAUUUAUGCAAAGCGAAACGCACUCA-CUCGCAAAACAAUGGCGAUUUUACAG------------GCGCUACAGUGACGCAACAGUGCGUUAA ....((((......(((((((((((((((....))))..(((...))).....-.((((.........))))))))))))------------)))......))))(((....)))..... ( -29.00) >DroSim_CAF1 4981 108 - 1 GAAAGUCAAUUAAACGCUUGUAAAAUGCAUUUAUGCAAAGCGAAACGCACUCUGGUCGCAAAACAAUGGCGAUUUUACAG------------GCGCUACAGUGACGCAACAGUGCGUUAA ....((((......((((((((((.((((....))))..(((...)))......(((((.........))))))))))))------------)))......))))(((....)))..... ( -29.40) >DroYak_CAF1 5153 120 - 1 GAAAGUCAAUUAAACGCUUGUAAAAUGCAUUUAUGCAAAGCAAAACGCACUCUGGUCGCACAACAAUGGCGAUUUCACAGUCGCUCGAUGUUGAGCUACAGUGACGCAACAGUGCGUUAA ...............((((......((((....))))))))..((((((((.(((((((.(((((.(((((((......))))).)).))))).(...).))))).))..)))))))).. ( -33.90) >consensus GAAAGUCAAUUAAACGCUUGUAAAAUGCAUUUAUGCAAAGCGAAACGCACUCUGGUCGCAAAACAAUGGCGAUUUUACAG____________GCGCUACAGUGACGCAACAGUGCGUUAA ..............(((((......((((....))))))))).((((((((....((((.........))))....................((((......).)))...)))))))).. (-22.21 = -22.52 + 0.31)

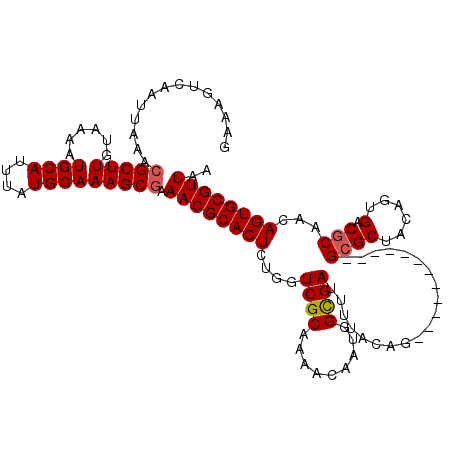
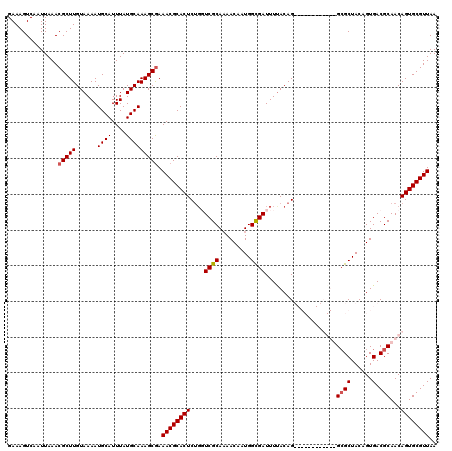
| Location | 5,226,762 – 5,226,868 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -26.83 |
| Energy contribution | -26.82 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5226762 106 + 22407834 AAAACGUGUCACAGGCAAUACAAACAUGUUUUUGCUGUUAGCCAGCAGCUGUAAGCGGCAUGAAUGG-------CCAACUGGCUAUGGCUUCGGUUUU-------UGCCUUCCGAUAAAU ......((((..((((((.(((....)))....((((..((((....((((....))))....((((-------((....)))))))))).))))..)-------)))))...))))... ( -31.30) >DroSec_CAF1 4211 106 + 1 AAAACGUGUCACAGGCAAUAAAAACAUGUUUUUGCUGUUAGCCAGCAGCUGUAAGCGGCAUGAAUGG-------CCAACUGGCUUCGGCUUCGGUUUU-------UGCCUUCCGAUAAAU ......((((..((((((...((((.((.....((((..((((((..((.....))(((.......)-------))..)))))).))))..)))))))-------)))))...))))... ( -31.10) >DroSim_CAF1 5089 106 + 1 AAAACGUGUCACAGGCAAUACAAACAUGUUUUUGCUGUUAGCCAGCAGCUGUAAGCGGCAUGAAUGG-------CCAACUGGCUUCGGCUUCGGUUUU-------UGCCUUCCGAUAAAU ......((((..((((((...((((.((.....((((..((((((..((.....))(((.......)-------))..)))))).))))..)))))))-------)))))...))))... ( -31.10) >DroYak_CAF1 5273 120 + 1 AAAACGUGUCACAGGCAAUACAAACAUGUUUUUGCUGUUAGCCAGCAGCUGUAAGCCGCAUGAAUGGCCAAUGGCCAACUGGCUCUGGCUUUGGCUAUUGCCUAUUGCCUUCCGAUAAAU ......((((..((((((((....(((((...(((((.....)))))((.....)).)))))...(((.(((((((((..(((....)))))))))))))))))))))))...))))... ( -41.70) >consensus AAAACGUGUCACAGGCAAUACAAACAUGUUUUUGCUGUUAGCCAGCAGCUGUAAGCGGCAUGAAUGG_______CCAACUGGCUUCGGCUUCGGUUUU_______UGCCUUCCGAUAAAU ......((((..(((((..........(((...((((..((((((..((((....)))).....((((.....)))).)))))).))))...)))..........)))))...))))... (-26.83 = -26.82 + -0.00)



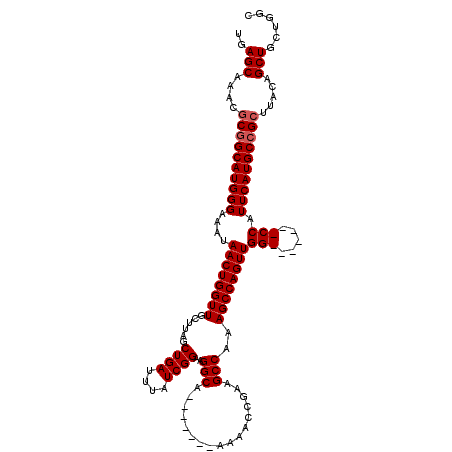
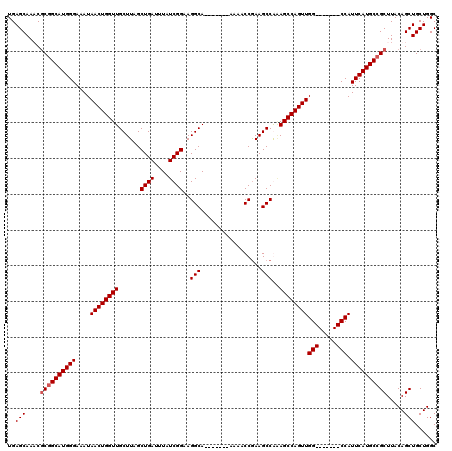
| Location | 5,226,802 – 5,226,908 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -31.72 |
| Energy contribution | -32.41 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5226802 106 - 22407834 UGAGCAAUCGCGGCAUGGGAAAUAACUGGUUGCUUAGCUGAUUUAUCGGAAGGCA-------AAAACCGAAGCCAUAGCCAGUUGG-------CCAUUCAUGCCGCUUACAGCUGCUGGC ..(((....((((((((((...((((((((((.....((((....))))..(((.-------.........))).)))))))))).-------...)))))))))).....)))...... ( -36.90) >DroSec_CAF1 4251 106 - 1 UGAGCAAACGCGGCAUGGGAAAUAACUGGUUGCUUAGCUGAUUUAUCGGAAGGCA-------AAAACCGAAGCCGAAGCCAGUUGG-------CCAUUCAUGCCGCUUACAGCUGCUGGC ..(((....((((((((((...(((((((((((...)).......((((..((..-------....))....))))))))))))).-------...)))))))))).....)))...... ( -36.40) >DroSim_CAF1 5129 106 - 1 UGAGCAAACGCGGCAUGGGAAAUAACUGGUUGCUUAGCUGAUUUAUCGGAAGGCA-------AAAACCGAAGCCGAAGCCAGUUGG-------CCAUUCAUGCCGCUUACAGCUGCUGGC ..(((....((((((((((...(((((((((((...)).......((((..((..-------....))....))))))))))))).-------...)))))))))).....)))...... ( -36.40) >DroYak_CAF1 5313 120 - 1 UGAGCAAUCUCGGCAUGGGAAAUAACUGGUUGCUUAGCUGAUUUAUCGGAAGGCAAUAGGCAAUAGCCAAAGCCAGAGCCAGUUGGCCAUUGGCCAUUCAUGCGGCUUACAGCUGCUGGC .(((....)))(((...((...(((((((((((((..((((....)))).))))....(((....)))........))))))))).))....)))..(((.(((((.....)))))))). ( -41.10) >consensus UGAGCAAACGCGGCAUGGGAAAUAACUGGUUGCUUAGCUGAUUUAUCGGAAGGCA_______AAAACCGAAGCCAAAGCCAGUUGG_______CCAUUCAUGCCGCUUACAGCUGCUGGC ..(((....((((((((((....((((((((......((((....))))..(((.................)))..))))))))(((.....))).)))))))))).....)))...... (-31.72 = -32.41 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:39 2006