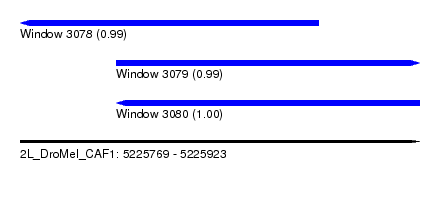
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,225,769 – 5,225,923 |
| Length | 154 |
| Max. P | 0.995490 |

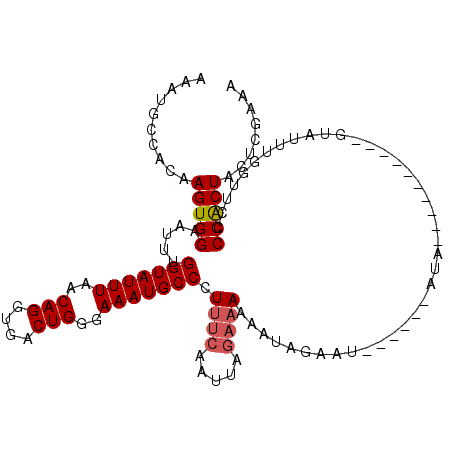
| Location | 5,225,769 – 5,225,884 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5225769 115 - 22407834 AAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUCAAAAAAAAAAAAUAGGAAUAUGAAAGCGCGGUCUAUUUGGUUCGCCACUGUUCGAAA ........((((((((....(((((((..(((....)))..)))))))((((((((((...............)))).)))))).....)))))))).((((........)))). ( -29.76) >DroSec_CAF1 3241 99 - 1 AAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUAGAAAAAAUAGAAU------AUA----------GUAUUUUGUUCCCCGCUACUCGAAA ..........(((((.....(((((((..(((....)))..))))))).((((.....)))).((((((((------(..----------.)))))))))..)))))........ ( -24.30) >DroSim_CAF1 4131 99 - 1 AAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUAGAAAAAAUAGAGU------AUA----------GUAUUUGGUUCCCCGCUACUCGAAA ..........(((((.....(((((((..(((....)))..)))))))..........(((.(((((....------...----------.)))))..))).)))))........ ( -23.20) >DroYak_CAF1 4285 111 - 1 AAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGAAAAUGCCCUGCCAAUAGGAAAAAAU----UAGAAAUAUGAAAAUGUAGUGUAUUUUGUUCCCCACUACUCAAAA ..........(((((.....(((((((..(((....)))..))))))).........(((((((((----.....((((....)))).....))))).)))))))))........ ( -27.50) >consensus AAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUAGAAAAAAUAGAAU______AUA__________GUAUUUGGUUCCCCACUACUCGAAA ..........(((((.....(((((((..(((....)))..))))))).((((.....))))........................................)))))........ (-19.88 = -20.38 + 0.50)

| Location | 5,225,806 – 5,225,923 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5225806 117 + 22407834 UAUUCCUAUUUUUUUUUUUUGAAUUGAAAGGGCAUUUCCCAGUCACCUGUUAAAUACCAAAUUCCACUUGUGGCAUUUGUGCUUUUCUGCACGUUUUCCUCCGUUCAAUUUU-UUUAU ....................((((((((.(((.........(((((..((..(((.....)))..))..)))))....((((......)))).......))).)))))))).-..... ( -21.10) >DroSec_CAF1 3268 112 + 1 U------AUUCUAUUUUUUCUAAUUGAAAGGGCAUUUCCCAGUCACCUGUUAAAUACCAAAUUCCACUUGUGGCAUUUGUGCUUUUCCGCACGUUUUCCUCCAUUCAAUUUUUUUUAU .------..............(((((((.(((.........(((((..((..(((.....)))..))..)))))....((((......)))).......))).)))))))........ ( -19.20) >DroSim_CAF1 4158 106 + 1 U------ACUCUAUUUUUUCUAAUUGAAAGGGCAUUUCCCAGUCACCUGUUAAAUACCAAAUUCCACUUGUGGCAUUUGUGCUUUUCCGCACGUUUUCCUCCAUUCAAUUUU------ .------..............(((((((.(((.........(((((..((..(((.....)))..))..)))))....((((......)))).......))).)))))))..------ ( -19.20) >DroYak_CAF1 4322 114 + 1 UAUUUCUA----AUUUUUUCCUAUUGGCAGGGCAUUUUCCAGUCACCUGUUAAAUACCAAAUUCCACUUGUGGCAUUUGUGCUUUUCUGCACGUUUUCCUCCGUUCAUUUUGUUUUUG ........----.............((.((((.........(((((..((..(((.....)))..))..)))))....((((......))))....))))))................ ( -18.60) >consensus U______AUUCUAUUUUUUCUAAUUGAAAGGGCAUUUCCCAGUCACCUGUUAAAUACCAAAUUCCACUUGUGGCAUUUGUGCUUUUCCGCACGUUUUCCUCCAUUCAAUUUU_UUUAU .....................(((((((.(((.........(((((..((..(((.....)))..))..)))))....((((......)))).......))).)))))))........ (-17.74 = -18.30 + 0.56)

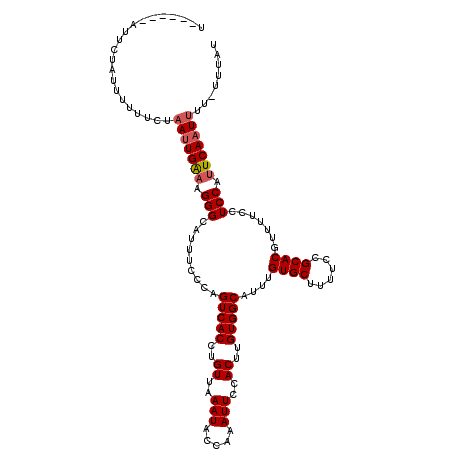
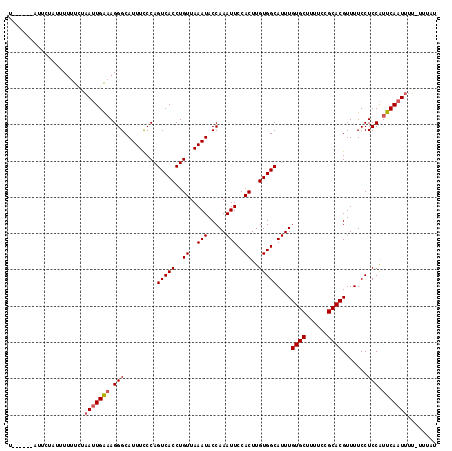
| Location | 5,225,806 – 5,225,923 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -5.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5225806 117 - 22407834 AUAAA-AAAAUUGAACGGAGGAAAACGUGCAGAAAAGCACAAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUCAAAAAAAAAAAAUAGGAAUA .....-....(((((.(((((.....((((......))))((((.(((....))).))))(((((((..(((....)))..))))))))))))..))))).................. ( -30.50) >DroSec_CAF1 3268 112 - 1 AUAAAAAAAAUUGAAUGGAGGAAAACGUGCGGAAAAGCACAAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUAGAAAAAAUAGAAU------A ..........((((.((((((.....((((......))))((((.(((....))).))))(((((((..(((....)))..))))))))))))).))))............------. ( -31.30) >DroSim_CAF1 4158 106 - 1 ------AAAAUUGAAUGGAGGAAAACGUGCGGAAAAGCACAAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUAGAAAAAAUAGAGU------A ------....((((.((((((.....((((......))))((((.(((....))).))))(((((((..(((....)))..))))))))))))).))))............------. ( -31.30) >DroYak_CAF1 4322 114 - 1 CAAAAACAAAAUGAACGGAGGAAAACGUGCAGAAAAGCACAAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGAAAAUGCCCUGCCAAUAGGAAAAAAU----UAGAAAUA ................((.((.....((((......))))((((.(((....))).))))(((((((..(((....)))..))))))))).)).............----........ ( -26.50) >consensus AUAAA_AAAAUUGAACGGAGGAAAACGUGCAGAAAAGCACAAAUGCCACAAGUGGAAUUUGGUAUUUAACAGGUGACUGGGAAAUGCCCUUUCAAUUAGAAAAAAUAGAAU______A ................(((((.....((((......))))((((.(((....))).))))(((((((..(((....)))..))))))))))))......................... (-28.56 = -28.62 + 0.06)

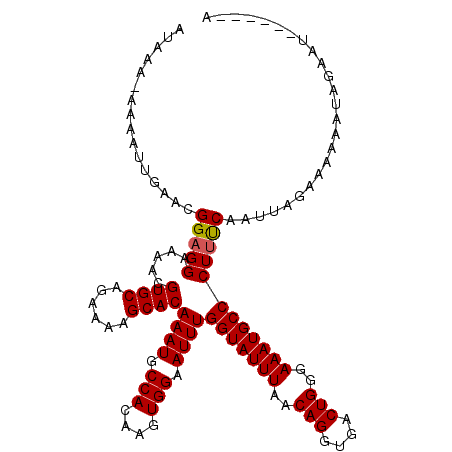
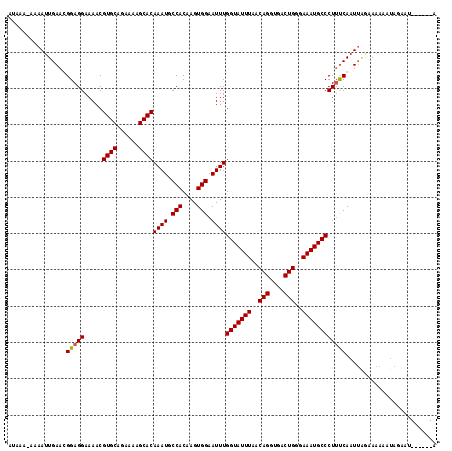
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:33 2006