
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,223,346 – 5,223,799 |
| Length | 453 |
| Max. P | 0.997733 |

| Location | 5,223,346 – 5,223,448 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -25.86 |
| Energy contribution | -25.36 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223346 102 - 22407834 GGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGUUUGCGUCUCGCUCUUUAGUAUUUGUAUUUGU------------AUCUGCAAC------UGAAAGUGCGCACUGUGA .((((((....((((((((((.((((((......))))))))))))))))...((.(((((((...((((.....------------...))))))------)).))).)))))).)).. ( -31.50) >DroSec_CAF1 839 102 - 1 GGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGCCUGGGUCUCGCUCUUUAGUAUUUGUAUUUGU------------AUCUGCAAC------UGAAAUUGCGCACUGUGA .((((((.(((((((((((((.((((((......))))))))))))(((.....)))....))))))).......------------....((((.------.....)))))))).)).. ( -32.30) >DroSim_CAF1 1700 102 - 1 GGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGCCUGGGUCUCGCUCUUUAGUAUUUGUAUUUGU------------AUCUGCAAC------UGAAACUGCGCACUGUGA .((((((....((.(((((((.((((((......))))))))))))).))...((..((((((...((((.....------------...))))))------))))...)))))).)).. ( -32.00) >DroYak_CAF1 1772 120 - 1 GGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGUUAGCGUCUUGCUCUUUAGUAUUUGUAUUUGUAUUUGAAUUUGUAUCUGCAACUGAAACUGAAACUGCGCACUGUGA .((((((....((((..((((.((((((......))))))))))..))))...((..((((((.((.((...((((..((......))..)))))).)).))))))...)))))).)).. ( -27.40) >consensus GGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGCCUGCGUCUCGCUCUUUAGUAUUUGUAUUUGU____________AUCUGCAAC______UGAAACUGCGCACUGUGA .((((((.(((((((((((((.((((((......)))))))))))........(((....)))...)))))))).................(((..............))))))).)).. (-25.86 = -25.36 + -0.50)

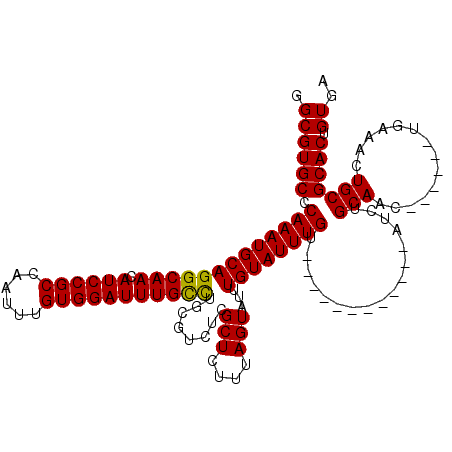
| Location | 5,223,373 – 5,223,488 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223373 115 - 22407834 UAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGUUUGCGUCUCGCUCUUUAGUAUUUGUAUUUGU----- (((((((..(((((......)))))......(.(((..((((....)))).((((((((((.((((((......))))))))))))))))...)))))))))))...........----- ( -29.10) >DroSec_CAF1 866 115 - 1 UAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUGGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGCCUGGGUCUCGCUCUUUAGUAUUUGUAUUUGU----- (((((((.....................((....))...(((((.(((((......(((((.((((((......))))))))))))))))..))))))))))))...........----- ( -32.50) >DroSim_CAF1 1727 115 - 1 UAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUGGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGCCUGGGUCUCGCUCUUUAGUAUUUGUAUUUGU----- (((((((.....................((....))...(((((.(((((......(((((.((((((......))))))))))))))))..))))))))))))...........----- ( -32.50) >DroYak_CAF1 1812 120 - 1 UAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGUUAGCGUCUUGCUCUUUAGUAUUUGUAUUUGUAUUUG (((((((..(((((......)))))......(.(((((((((....)))).((((..((((.((((((......))))))))))..)))).)))))))))))))................ ( -26.90) >consensus UAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAUUUGCCUGCGUCUCGCUCUUUAGUAUUUGUAUUUGU_____ (((((((..((.................((....))..((((....)))).((((((((((.((((((......))))))))))))))))...))..)))))))................ (-28.70 = -28.95 + 0.25)


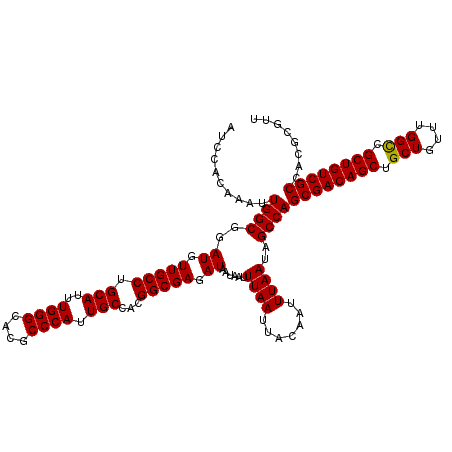
| Location | 5,223,408 – 5,223,528 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -41.35 |
| Consensus MFE | -41.54 |
| Energy contribution | -41.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.40 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223408 120 + 22407834 AUCCACAAAUUGGCGGAUGUUGCCUGCAUUUGGGCACGCCCAUUGCUACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGUCGCUGUCGCCACGCGUU ..........((((..((.(((((.(((..((((....)))).)))...))))).))......((((.......))))..))))((((((((.(((....))).))))))))........ ( -40.60) >DroSec_CAF1 901 120 + 1 AUCCACAAAUUGGCGGAUGUUGCCUGCAUUUGGGCACGCCCAUUGCCACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUU ..........((((..((.(((((.(((..((((....)))).)))...))))).))......((((.......))))..))))((((((((.(((....))).))))))))........ ( -41.50) >DroSim_CAF1 1762 120 + 1 AUCCACAAAUUGGCGGAUGUUGCCUGCAUUUGGGCACGCCCAUUGCCACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUU ..........((((..((.(((((.(((..((((....)))).)))...))))).))......((((.......))))..))))((((((((.(((....))).))))))))........ ( -41.50) >DroYak_CAF1 1852 120 + 1 AUCCACAAAUUGGCGGAUGUUGCCUGCAUUUGGGCACGCCCAUUGCUACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUGGGCCGCUGUCGCCACGCGUU ..........((((..((.(((((.(((..((((....)))).)))...))))).))......((((.......))))..))))((((((((.(((....))).))))))))........ ( -41.80) >consensus AUCCACAAAUUGGCGGAUGUUGCCUGCAUUUGGGCACGCCCAUUGCCACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUU ..........((((..((.(((((.(((..((((....)))).)))...))))).))......((((.......))))..))))((((((((.(((....))).))))))))........ (-41.54 = -41.35 + -0.19)

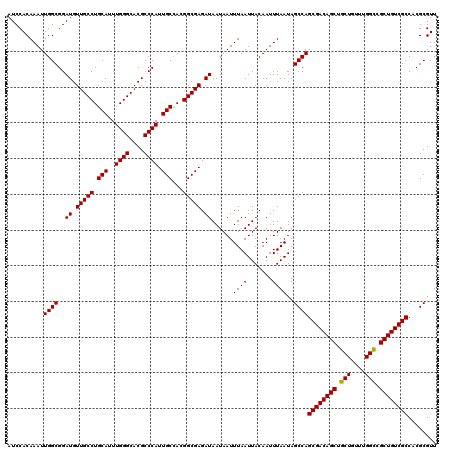
| Location | 5,223,408 – 5,223,528 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -40.97 |
| Energy contribution | -41.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.939994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223408 120 - 22407834 AACGCGUGGCGACAGCGACCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAU ...(((((((((((((..(......)..))))))))).(((((......(((((......)))))......)))))..((((....)))).)))).(....)((((((......)))))) ( -39.30) >DroSec_CAF1 901 120 - 1 AACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUGGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAU ...((.((((((((((.((......)).))))))))))))............................(((...(((.((((....))))..))).)))...((((((......)))))) ( -42.80) >DroSim_CAF1 1762 120 - 1 AACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUGGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAU ...((.((((((((((.((......)).))))))))))))............................(((...(((.((((....))))..))).)))...((((((......)))))) ( -42.80) >DroYak_CAF1 1852 120 - 1 AACGCGUGGCGACAGCGGCCCAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAU ...(((((((((((((.((......)).))))))))).(((((......(((((......)))))......)))))..((((....)))).)))).(....)((((((......)))))) ( -41.90) >consensus AACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUGGGCGUGCCCAAAUGCAGGCAACAUCCGCCAAUUUGUGGAU ...((.((((((((((.((......)).))))))))))))............................(((...(((.((((....))))..))).)))...((((((......)))))) (-40.97 = -41.23 + 0.25)

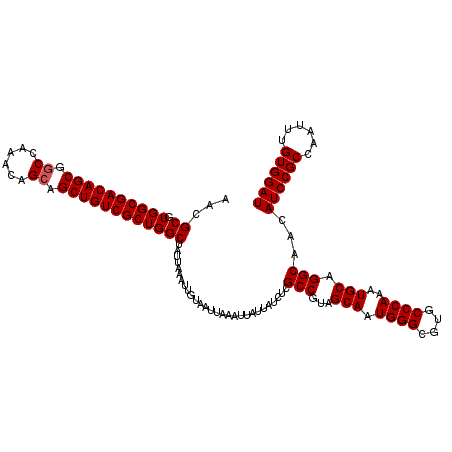
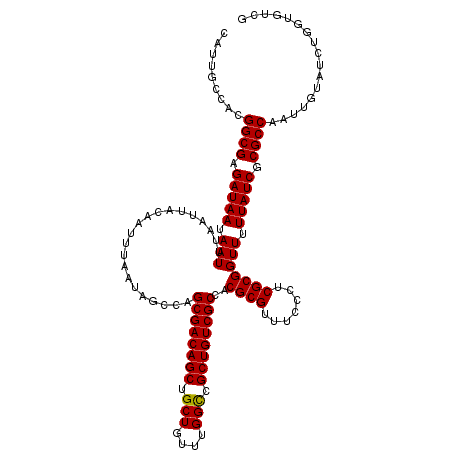
| Location | 5,223,448 – 5,223,568 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -37.59 |
| Energy contribution | -37.40 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223448 120 + 22407834 CAUUGCUACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGUCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGAUGUCG ..((((....))))((((.....(((..(((((((.....((..((((((((.(((....))).))))))))...))........((((((....))))))..)))))))..))))))). ( -38.10) >DroSec_CAF1 941 120 + 1 CAUUGCCACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCG ..((((....))))(((((.(((.....................((((((((.(((....))).))))))))..((((.......))))))).)))))((((((........)))))).. ( -39.20) >DroSim_CAF1 1802 120 + 1 CAUUGCCACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCG ..((((....))))(((((.(((.....................((((((((.(((....))).))))))))..((((.......))))))).)))))((((((........)))))).. ( -39.20) >DroYak_CAF1 1892 120 + 1 CAUUGCUACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUGGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGCUGCUG .........((((.(((((.(((.....................((((((((.(((....))).))))))))..((((.......))))))).))))).))))................. ( -37.50) >consensus CAUUGCCACGGCGAGAUAAUAAUUUAAUUACAAUUUAAUAGCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCG .........((((.(((((.(((.....................((((((((.(((....))).))))))))..((((.......))))))).))))).))))................. (-37.59 = -37.40 + -0.19)

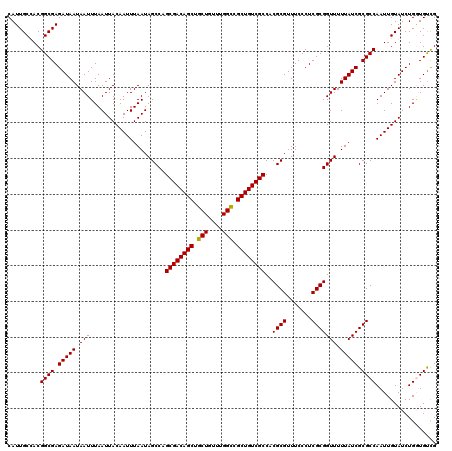
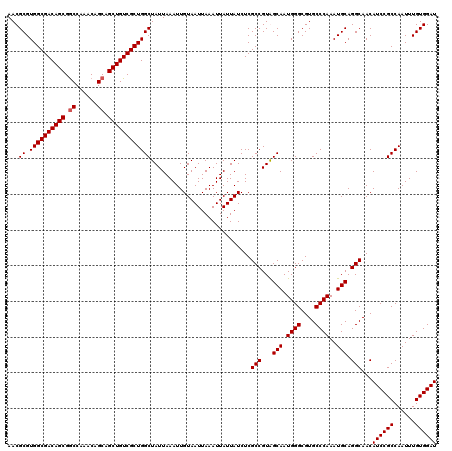
| Location | 5,223,448 – 5,223,568 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -37.03 |
| Energy contribution | -37.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223448 120 - 22407834 CGACAUCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGACCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUG .................((((.(((((...((....)).....((.((((((((((..(......)..)))))))))))).....................))))).))))......... ( -35.70) >DroSec_CAF1 941 120 - 1 CGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUGGCAAUG .....(((........(((((.(((((...((....)).....((.((((((((((.((......)).)))))))))))).....................))))).))))))))..... ( -38.60) >DroSim_CAF1 1802 120 - 1 CGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUGGCAAUG .....(((........(((((.(((((...((....)).....((.((((((((((.((......)).)))))))))))).....................))))).))))))))..... ( -38.60) >DroYak_CAF1 1892 120 - 1 CAGCAGCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCCAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUG ..((.((.(((...)))((((.(((((...((....)).....((.((((((((((.((......)).)))))))))))).....................))))).)))))).)).... ( -39.10) >consensus CGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGCUAUUAAAUUGUAAUUAAAUUAUUAUCUCGCCGUAGCAAUG .................((((.(((((...((....)).....((.((((((((((.((......)).)))))))))))).....................))))).))))......... (-37.03 = -37.27 + 0.25)

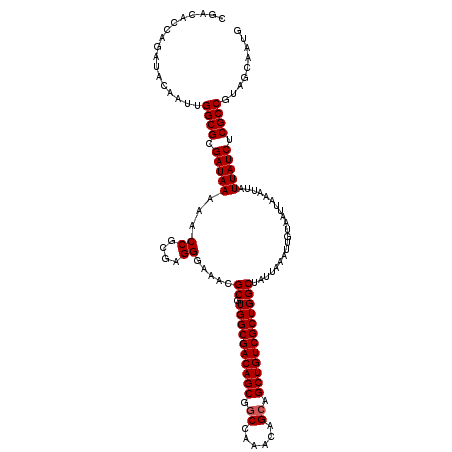
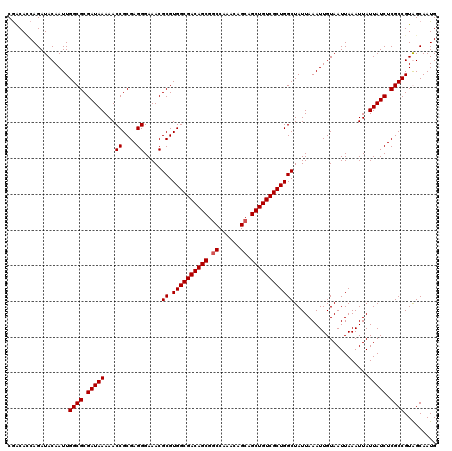
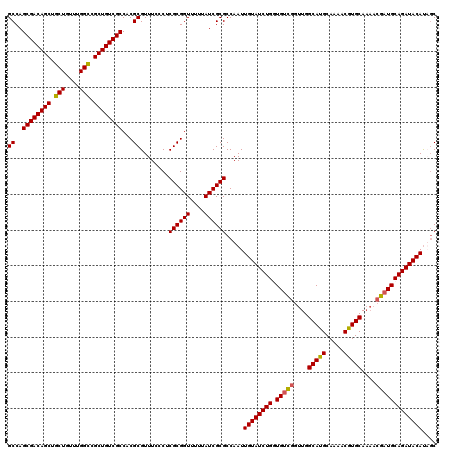
| Location | 5,223,488 – 5,223,608 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -49.10 |
| Consensus MFE | -47.61 |
| Energy contribution | -47.55 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223488 120 + 22407834 GCCAGCGACAGCUGCUGUUUGGUCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGAUGUCGGUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACAUAGC ((..((((((((.(((....))).))))))))...))........((((((....)))))).....((((((((.(((((....(((((.....)))))....))))))))))))).... ( -48.10) >DroSec_CAF1 981 120 + 1 GCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCGGUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACACUGC ((..((((((((.(((....))).))))))))...))........((((((....)))))).....((((((((.(((((....(((((.....)))))....))))))))))))).... ( -49.60) >DroSim_CAF1 1842 120 + 1 GCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCGGUUGGCAUGCAAAACAUGCAAAACGAUGCAGAUACAUUGC ((..((((((((.(((....))).))))))))...))........((((((....)))))).((((.(((((((.(((((....(((((.....)))))....)))))))))))))))). ( -50.80) >DroYak_CAF1 1932 116 + 1 GCCAGCGACAGCUGCUGUUGGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGCUGCUGUUUGGCAUGCAAAGCGUGCA----GAUGCAGAUACAUACC ((..((((((((.(((....))).))))))))...))........((((((....)))))).....(((((((((....(((((.(((((...)))))))----)))))))))))).... ( -47.90) >consensus GCCAGCGACAGCUGCUGUUUGGCCGCUGUCGCCACGCGUUUCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCGGUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACAUAGC ((..((((((((.(((....))).))))))))...))........((((((....)))))).....((((((((.(((((....(((((.....)))))....))))))))))))).... (-47.61 = -47.55 + -0.06)

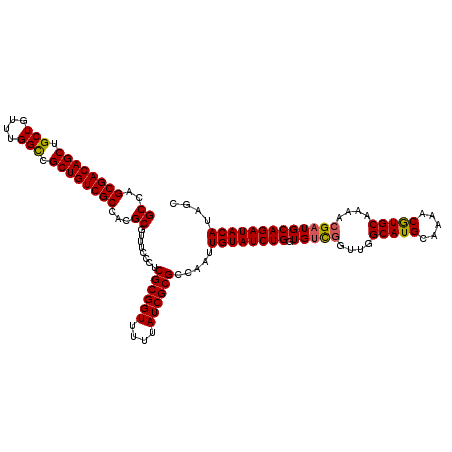
| Location | 5,223,488 – 5,223,608 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -47.28 |
| Consensus MFE | -41.75 |
| Energy contribution | -42.62 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
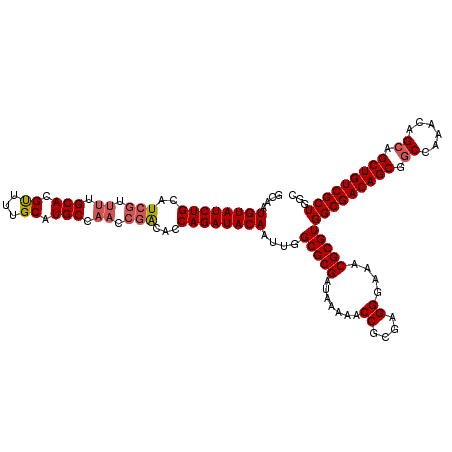
>2L_DroMel_CAF1 5223488 120 - 22407834 GCUAUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAACCGACAUCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGACCAAACAGCAGCUGUCGCUGGC (((.((((((((..(((.((.(((.(.....).))).)).)))...))))))))....(((((.......((....))....)))))(((((((((..(......)..)))))))))))) ( -43.50) >DroSec_CAF1 981 120 - 1 GCAGUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAACCGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGC .(((((((((((..(((.((.(((.(.....).))).)).)))...))))))).))))(((((.......((....))....)))))(((((((((.((......)).)))))))))... ( -46.40) >DroSim_CAF1 1842 120 - 1 GCAAUGUAUCUGCAUCGUUUUGCAUGUUUUGCAUGCCAACCGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGC .(((((((((((..(((.((.(((((.....))))).)).)))...))))))).))))(((((.......((....))....)))))(((((((((.((......)).)))))))))... ( -50.70) >DroYak_CAF1 1932 116 - 1 GGUAUGUAUCUGCAUC----UGCACGCUUUGCAUGCCAAACAGCAGCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCCAACAGCAGCUGUCGCUGGC ....(((((((((...----((((.....))))(((......))))))))))))....(((((.......((....))....)))))(((((((((.((......)).)))))))))... ( -48.50) >consensus GCAAUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAACCGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGAAACGCGUGGCGACAGCGGCCAAACAGCAGCUGUCGCUGGC ....((((((((..(((.((.(((.((...)).))).)).)))...))))))))....(((((.......((....))....)))))(((((((((.((......)).)))))))))... (-41.75 = -42.62 + 0.88)

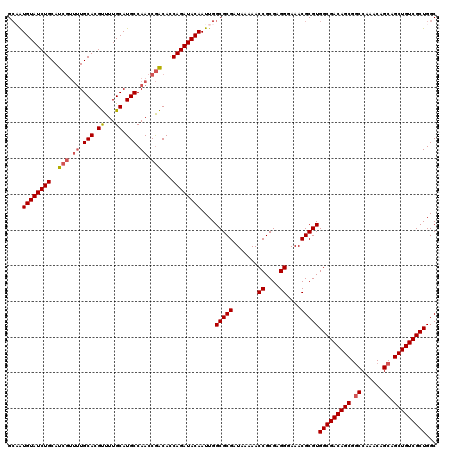
| Location | 5,223,528 – 5,223,648 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -31.75 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223528 120 + 22407834 UCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGAUGUCGGUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACAUAGCCAGCCAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUG ......(((((....)))))((....((((((((.(((((....(((((.....)))))....)))))))))))))..))((((...((((.....))))((((......))))..)))) ( -36.60) >DroSec_CAF1 1021 120 + 1 UCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCGGUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACACUGCCAGCCAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUG .....((((((....)))))).....((((((((.(((((....(((((.....)))))....)))))))))))))....((((...((((.....))))((((......))))..)))) ( -36.20) >DroSim_CAF1 1882 120 + 1 UCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCGGUUGGCAUGCAAAACAUGCAAAACGAUGCAGAUACAUUGCCAGCAAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUG .....((((((....)))))).((((.(((((((.(((((....(((((.....)))))....)))))))))))))))).((((...((((.....))))((((......))))..)))) ( -38.30) >DroYak_CAF1 1972 112 + 1 UCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGCUGCUGUUUGGCAUGCAAAGCGUGCA----GAUGCAGAUACAUACCC----ACCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUG .....((((((....)))))).(((((((((((((....(((((.(((((...)))))))----)))))))))))).....----((((((.....))))))...))))........... ( -35.30) >consensus UCCCUCGCGGUUUUUAUCGCGCCAAUUGUAUCUGGUGUCGGUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACAUAGCCAGCCAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUG .....((((((....)))))).....((((((((.(((((....(((((.....)))))....)))))))))))))....((((...((((.....))))((((......))))..)))) (-31.75 = -32.62 + 0.88)

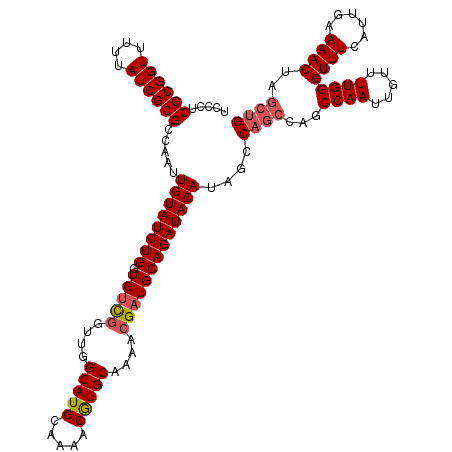
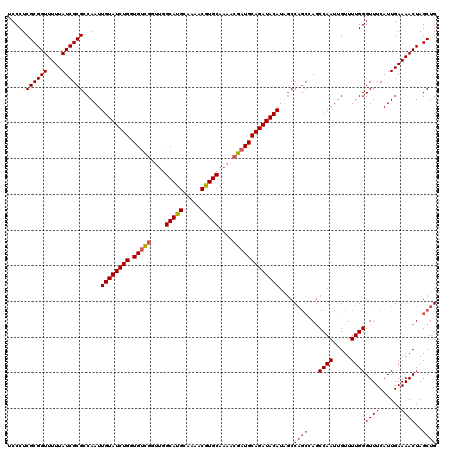
| Location | 5,223,528 – 5,223,648 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223528 120 - 22407834 CAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUGGCUGGCUAUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAACCGACAUCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGA ((((((((...((((.............))))))))))))((((((((((((..(((.((.(((.(.....).))).)).)))...))))))))..))))..........((....)).. ( -35.02) >DroSec_CAF1 1021 120 - 1 CAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUGGCUGGCAGUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAACCGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGA ...................(((....((((((.((((.((((((.....)))).(((.((.(((.(.....).))).)).)))))))))...)))))).((((........)))).))). ( -34.50) >DroSim_CAF1 1882 120 - 1 CAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUUGCUGGCAAUGUAUCUGCAUCGUUUUGCAUGUUUUGCAUGCCAACCGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGA ..((((((..(((((.............)))))...))))))..((((((((..(((.((.(((((.....))))).)).)))...)))))))).....((((........))))..... ( -35.32) >DroYak_CAF1 1972 112 - 1 CAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGGU----GGGUAUGUAUCUGCAUC----UGCACGCUUUGCAUGCCAAACAGCAGCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGA ..(((((((....(((..((((((.....))))))----...)))((((((((...----((((.....))))(((......))))))))))))))))))..........((....)).. ( -36.10) >consensus CAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUGGCUGGCAAUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAACCGACACCAGAUACAAUUGGCGCGAUAAAAACCGCGAGGGA ...................(((....((((((..(((.((((.(((((.(((((......)))).)...))))))))).))).(....)...)))))).((((........)))).))). (-26.02 = -26.90 + 0.88)

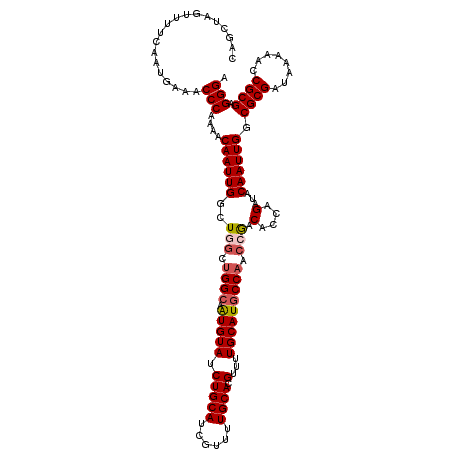
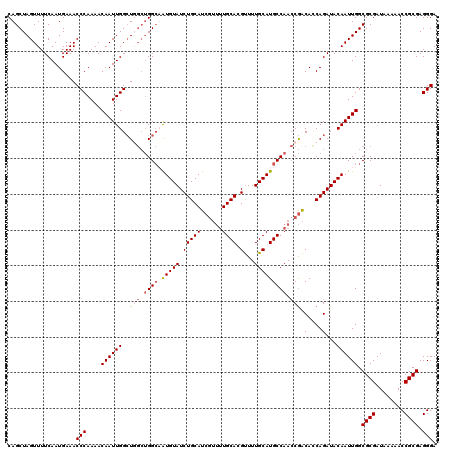
| Location | 5,223,568 – 5,223,686 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -28.94 |
| Energy contribution | -30.00 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223568 118 + 22407834 GUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACAUAGCCAGCCAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUGUGUUUC--AGCUAGCCAAGUUUUAUUCGCUGCCAACUUUU (((((((.(((((((.((((......))))((.(((((((.......((((.....))))((((......))))..))))))).))--..........)))))....))))))))).... ( -32.70) >DroSec_CAF1 1061 120 + 1 GUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACACUGCCAGCCAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUGUGCUUCACAGCUAGCCAAGUUUUAUUCGCUGCCAACUUUU (((((((.(((((((...(((((((((((((.....))))..........)))))))))((((((...)))).(((((((((...)))))))))))..)))))....))))))))).... ( -37.80) >DroSim_CAF1 1922 120 + 1 GUUGGCAUGCAAAACAUGCAAAACGAUGCAGAUACAUUGCCAGCAAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUGUGUUUCACAGCUAGCCAAGUUUUAUUCGCUGCCAACUUUU (((((((.(((((((........(((((.((((...(((....))).((((.....)))))))))))))....(((((((((...)))))))))....)))))....))))))))).... ( -36.70) >DroYak_CAF1 2012 110 + 1 UUUGGCAUGCAAAGCGUGCA----GAUGCAGAUACAUACCC----ACCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUGUGUUUC--AGCUAGCCAAGUUUUAUUCGCUGCCAACUUGU .((((((.((...((((...----.)))).((..((.....----((((((.....)))))).....))....(((((((.....)--))))))...........))))))))))..... ( -32.00) >consensus GUUGGCAUGCAAAACGUGCAAAACGAUGCAGAUACAUAGCCAGCCAGCCAAUUGUUUUGGGUUUCAUUGAAAACUAGCUGUGUUUC__AGCUAGCCAAGUUUUAUUCGCUGCCAACUUUU (((((((.((.((((.((((......)))).................((((.....)))))))).........(((((((((...))))))))).............))))))))).... (-28.94 = -30.00 + 1.06)

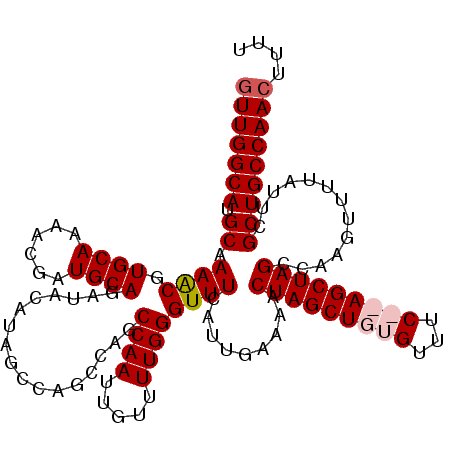
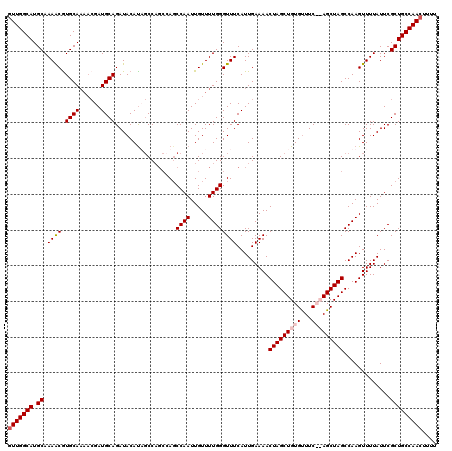
| Location | 5,223,568 – 5,223,686 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -29.17 |
| Energy contribution | -30.43 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223568 118 - 22407834 AAAAGUUGGCAGCGAAUAAAACUUGGCUAGCU--GAAACACAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUGGCUGGCUAUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAAC ....((((((((((((....((.((((((((.--......(((((((((.....((.....))....))))))))))))))))).))...((((......))))...))))).))))))) ( -38.01) >DroSec_CAF1 1061 120 - 1 AAAAGUUGGCAGCGAAUAAAACUUGGCUAGCUGUGAAGCACAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUGGCUGGCAGUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAAC ....(((((((((...........(((((((((((...)))))))))))......(((((.((((((....((....)).((((.....))))...))))))...))))))).))))))) ( -39.40) >DroSim_CAF1 1922 120 - 1 AAAAGUUGGCAGCGAAUAAAACUUGGCUAGCUGUGAAACACAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUUGCUGGCAAUGUAUCUGCAUCGUUUUGCAUGUUUUGCAUGCCAAC ....(((((((((((.........(((((((((((...)))))))))))...........((((.....)))).))))).((((...((.((((......)))).)).))))..)))))) ( -38.30) >DroYak_CAF1 2012 110 - 1 ACAAGUUGGCAGCGAAUAAAACUUGGCUAGCU--GAAACACAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGGU----GGGUAUGUAUCUGCAUC----UGCACGCUUUGCAUGCCAAA .((((((............))))))(((((((--(.....))))))))..........((((((.....))))))----.((((((((...((...----.....))..))))))))... ( -35.70) >consensus AAAAGUUGGCAGCGAAUAAAACUUGGCUAGCU__GAAACACAGCUAGUUUUCAAUGAAACCCAAAACAAUUGGCUGGCUGGCAAUGUAUCUGCAUCGUUUUGCACGUUUUGCAUGCCAAC ....((((((((((((........(((((((((((...)))))))))))...........((((.....)))).................((((......))))...))))).))))))) (-29.17 = -30.43 + 1.25)

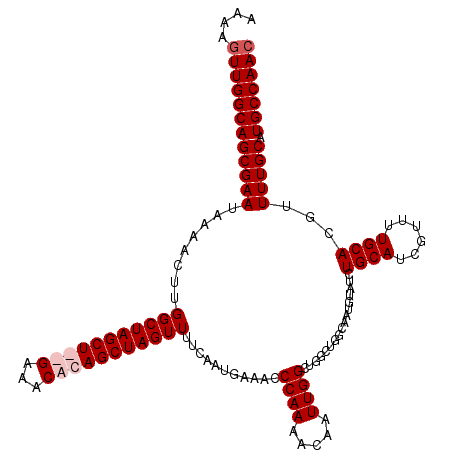
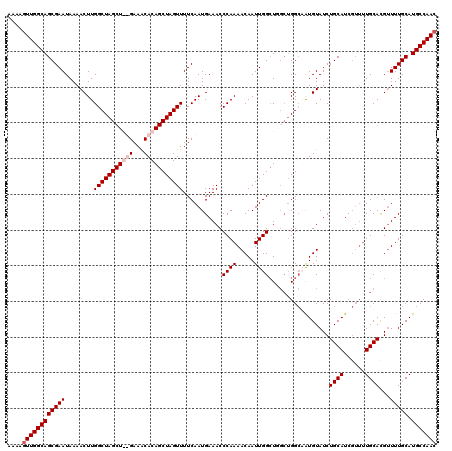
| Location | 5,223,686 – 5,223,799 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -24.79 |
| Energy contribution | -25.23 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5223686 113 + 22407834 AUCCGCAAAUACCUGUUGCUUCUCAUCUCUUUCAGCAAAAGCAACGAAAACAAAUCAC-------UGAGCACCGGGAACGAUGACCACGUAGCACGGUAGCCUGUUGCGGUGCAGACAAA ..((((((.((((((((((...(((((.......((....))............((.(-------((.....)))))..)))))....)))))).)))).....)))))).......... ( -28.00) >DroSec_CAF1 1181 113 + 1 AUCCGCAAAUACCUGUUGCUUCUCAUCUCCCUCAGCAAAAGCAACGAAAACAAAUCAC-------UGAGAACCGAGAACGAUGACCACGUAGCACGGUAGCCUGUUGCGGUGCAGACAAA ..((((((.((((((((((...(((((...(((.((....))............((..-------...))...)))...)))))....)))))).)))).....)))))).......... ( -28.00) >DroSim_CAF1 2042 113 + 1 AUCCGCAAAUACCUGUUGCUUCUCAUCUCUCUCAGCAAAAGCAAGGAAAACAAAUCAC-------UGAGAACCGAGAACGAUGACCACGUAGCACGGUAGCCUGUUGUGGUGCAGACAAA ..((((((.((((((((((...(((((..((((.((....))..((....((......-------))....))))))..)))))....)))))).)))).....)))))).......... ( -29.80) >DroYak_CAF1 2122 120 + 1 AUCCGCAAAUACCUGUUGCUUCUCAUCUCUUGCAGCAAAGCGAAAGAAAACAAAUCACAGAGAACUGAGAACUGAGAACGAUGACCACGUAGCACGGUAGCCUUUUGCGGUGCAGACAAA ..((((((((((((((((((((((((((.((((......)))).))).......((.(((....))).))..)))))).(.....)..)))))).))))....))))))).......... ( -31.20) >consensus AUCCGCAAAUACCUGUUGCUUCUCAUCUCUCUCAGCAAAAGCAACGAAAACAAAUCAC_______UGAGAACCGAGAACGAUGACCACGUAGCACGGUAGCCUGUUGCGGUGCAGACAAA ..((((((.((((((((((...(((((..((((.((....))............(((........))).....))))..)))))....)))))).)))).....)))))).......... (-24.79 = -25.23 + 0.44)

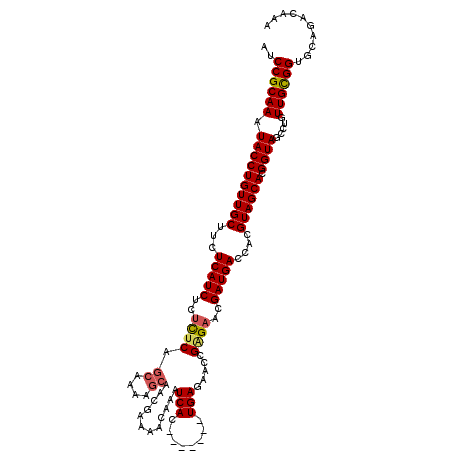
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:30 2006