| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 574,713 – 574,829 |
| Length | 116 |
| Max. P | 0.572905 |

| Location | 574,713 – 574,829 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -23.63 |
| Energy contribution | -23.96 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 574713 116 + 22407834 GAAGCCCGACUUU---GGGCCACACCUUUAAAAUA-UUUAGAACCUCUCGAAUAAAUAACAUAUUUUUAGUAAAUUUCGUGGCCGGUCUGGAGUGCUAUGUCUGCUCCAACCAGACGGGC ...(((((.....---.((((((..........((-(((((.....)).))))).......(((.....)))......))))))(((.((((((.........)))))))))...))))) ( -33.60) >DroPse_CAF1 14843 109 + 1 UAUGCUAGAGAUUUACGGGCCACACCUCAAUAAUACUUUAUAAUCUCCCCGAUA-----------UCUAGUGAGUUUUGUGGCCGGCCUGGAGUGCUAUGUGUGCUCCAACCAGACGGGC ...(((...........(((((((.((((((((....)))).(((.....))).-----------.....))))...)))))))((..(((((..(.....)..))))).)).....))) ( -31.80) >DroPer_CAF1 14827 109 + 1 UAUGCUAGAGAUUUACGGGCCACACCUCAAUAAUACUUUAUAAUCUCCCCGAUA-----------UCUAGUGAGUUUUGUGGCCGGCCUGGAGUGCUAUGUGUGCUCCAACCAGACGGGC ...(((...........(((((((.((((((((....)))).(((.....))).-----------.....))))...)))))))((..(((((..(.....)..))))).)).....))) ( -31.80) >consensus UAUGCUAGAGAUUUACGGGCCACACCUCAAUAAUACUUUAUAAUCUCCCCGAUA___________UCUAGUGAGUUUUGUGGCCGGCCUGGAGUGCUAUGUGUGCUCCAACCAGACGGGC ...(((.(.........(((((((.(((...........................................)))...)))))))((..((((((((.....)))))))).))...).))) (-23.63 = -23.96 + 0.34)

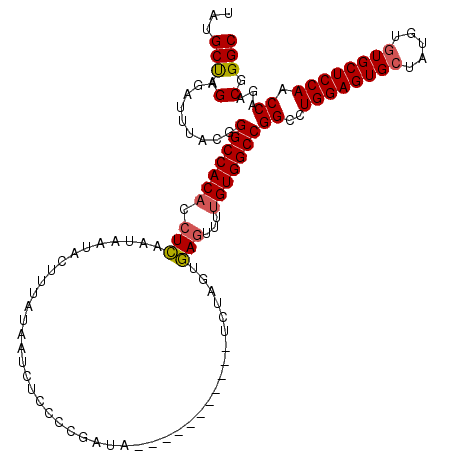
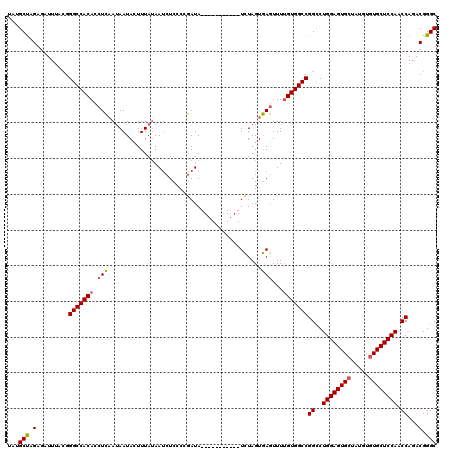
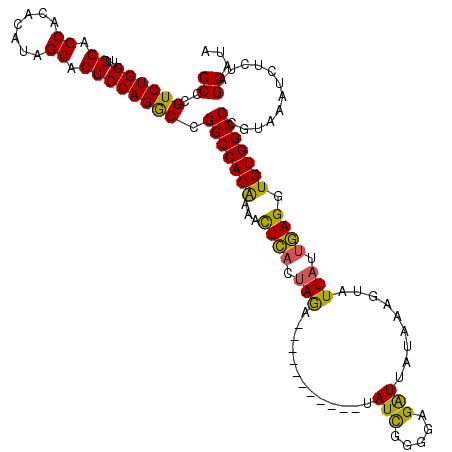
| Location | 574,713 – 574,829 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -26.01 |
| Energy contribution | -24.92 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 574713 116 - 22407834 GCCCGUCUGGUUGGAGCAGACAUAGCACUCCAGACCGGCCACGAAAUUUACUAAAAAUAUGUUAUUUAUUCGAGAGGUUCUAAA-UAUUUUAAAGGUGUGGCCC---AAAGUCGGGCUUC (((((.((((((((((...........))))).)))(((((((...((((....((((((.........((....))......)-)))))))))..))))))).---..)).)))))... ( -36.16) >DroPse_CAF1 14843 109 - 1 GCCCGUCUGGUUGGAGCACACAUAGCACUCCAGGCCGGCCACAAAACUCACUAGA-----------UAUCGGGGAGAUUAUAAAGUAUUAUUGAGGUGUGGCCCGUAAAUCUCUAGCAUA ((..((((((..((.((.......)).)))))))).(((((((...((((...((-----------(((...............)))))..)))).)))))))............))... ( -29.06) >DroPer_CAF1 14827 109 - 1 GCCCGUCUGGUUGGAGCACACAUAGCACUCCAGGCCGGCCACAAAACUCACUAGA-----------UAUCGGGGAGAUUAUAAAGUAUUAUUGAGGUGUGGCCCGUAAAUCUCUAGCAUA ((..((((((..((.((.......)).)))))))).(((((((...((((...((-----------(((...............)))))..)))).)))))))............))... ( -29.06) >consensus GCCCGUCUGGUUGGAGCACACAUAGCACUCCAGGCCGGCCACAAAACUCACUAGA___________UAUCGGGGAGAUUAUAAAGUAUUAUUGAGGUGUGGCCCGUAAAUCUCUAGCAUA ((..((((((..((.((.......)).)))))))).(((((((...((((.(((.............(((.....))).........))).)))).)))))))............))... (-26.01 = -24.92 + -1.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:28:51 2006