
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,216,041 – 5,216,201 |
| Length | 160 |
| Max. P | 0.940171 |

| Location | 5,216,041 – 5,216,161 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -32.35 |
| Energy contribution | -33.23 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5216041 120 + 22407834 AGCUACGUAUUUGGCAUUGUGCACCGCUUCCGUAUAGCAGGGUUCAUAGGCUAGUUUUGGAUAAUCCAAGGAUAUCAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGGUAUU ((((((((.....((.....)).((((((((.....((((((((((...(.((.((((((.....)))))).)).)....)).))))))))....)))..)))))))))))))....... ( -41.80) >DroSec_CAF1 5282 120 + 1 AGCUACGUAUUUGGCAUUGUGAACCGCUUCCAUAUAGCACGGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAUUAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGAUAUU ((((((((...((((.((((((((((((.......)))..)))))))))))))((((..((((.....(((((((......)).))))).))))..)))).....))))))))....... ( -38.70) >DroSim_CAF1 5302 120 + 1 AGCUACGUAUUUGGCAUUGUGCACCGCUUCCGUAUAGCAGGGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAUCAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGAUAUU ((((((((.....((.....)).((((((((.....((((((((((...(.((.((((((.....)))))).)).)....)).))))))))....)))..)))))))))))))....... ( -41.80) >DroYak_CAF1 11511 120 + 1 ACCUACAUAUUUGGCAUUGUGAACCGCUUCCGUAUAGCACGGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAACACAUUAUAAUCCUGUUUCAAGAACAGCGGGCGUAGCUGAAUAUU ......(((((..((.((((((((((((.......)))..)))))))))(((..(((((((.......(((((...........)))))..)))))))..))).......))..))))). ( -32.30) >consensus AGCUACGUAUUUGGCAUUGUGAACCGCUUCCGUAUAGCACGGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAUCAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGAUAUU ((((((((...((((.((((((((((((.......)))..)))))))))))))(((((.((((.....(((((...........))))).)))).))))).....))))))))....... (-32.35 = -33.23 + 0.88)

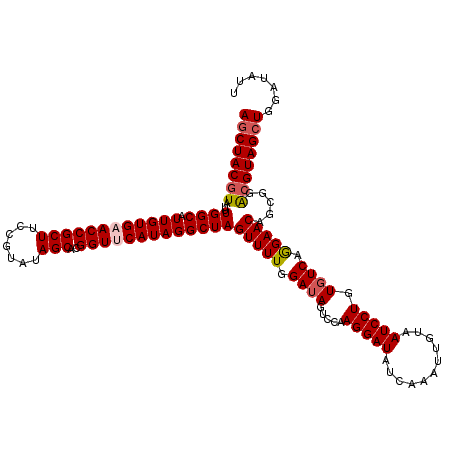
| Location | 5,216,041 – 5,216,161 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -25.81 |
| Energy contribution | -25.93 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5216041 120 - 22407834 AAUACCCAGCUACGUCCGCUGUUCCUGACACAGGAUUACAAUUUGAUAUCCUUGGAUUAUCCAAAACUAGCCUAUGAACCCUGCUAUACGGAAGCGGUGCACAAUGCCAAAUACGUAGCU .......(((((((((((((..(((((...))))).............(((((((.....))))...((((...........))))...)))))))).((.....)).....)))))))) ( -31.00) >DroSec_CAF1 5282 120 - 1 AAUAUCCAGCUACGUCCGCUGUUCCUGACACAGGAUUACAAUUUAAUAUCCUUGGACUAUCCAAAACUAGCCUAUGAACCGUGCUAUAUGGAAGCGGUUCACAAUGCCAAAUACGUAGCU .......((((((((..((((.(((((...)))))................((((.....))))...))))...((((((((.((....))..))))))))...........)))))))) ( -32.20) >DroSim_CAF1 5302 120 - 1 AAUAUCCAGCUACGUCCGCUGUUCCUGACACAGGAUUACAAUUUGAUAUCCUUGGACUAUCCAAAACUAGCCUAUGAACCCUGCUAUACGGAAGCGGUGCACAAUGCCAAAUACGUAGCU .......(((((((((((((..(((((...))))).............(((((((.....))))...((((...........))))...)))))))).((.....)).....)))))))) ( -31.00) >DroYak_CAF1 11511 120 - 1 AAUAUUCAGCUACGCCCGCUGUUCUUGAAACAGGAUUAUAAUGUGUUAUCCUUGGACUAUCCAAAACUAGCCUAUGAACCGUGCUAUACGGAAGCGGUUCACAAUGCCAAAUAUGUAGGU .........(((((...((((((.........((((.((.....)).))))((((.....))))))).)))...((((((((.((....))..))))))))............))))).. ( -23.80) >consensus AAUAUCCAGCUACGUCCGCUGUUCCUGACACAGGAUUACAAUUUGAUAUCCUUGGACUAUCCAAAACUAGCCUAUGAACCCUGCUAUACGGAAGCGGUGCACAAUGCCAAAUACGUAGCU .......(((((((((((((..(((((...))))).............(((((((.....))))...((((...........))))...))))))))...............)))))))) (-25.81 = -25.93 + 0.13)


| Location | 5,216,081 – 5,216,201 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -31.74 |
| Energy contribution | -32.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5216081 120 + 22407834 GGUUCAUAGGCUAGUUUUGGAUAAUCCAAGGAUAUCAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGGUAUUGGGACUGAAAUCGCGGUGCCCGUUGAAUCCAUGGAUCAGA (((((((..(((.((((..((((.....(((((..(.....)..))))).))))..)))))))(((..((((.((((((((.((......)).))))))))))))..))))))))))... ( -42.30) >DroSec_CAF1 5322 120 + 1 GGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAUUAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGAUAUUGGGACUGAAAUCGCGGUGCCCGUUGAAUCCAUGGAUCAGA (((((((..(((.((((..((((.....(((((((......)).))))).))))..)))))))(((..((((.((.(((((.((......)).))))))).))))..))))))))))... ( -35.30) >DroSim_CAF1 5342 120 + 1 GGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAUCAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGAUAUUGGGACUGAAAUCGCGGUGCCCGUUGAAUCCAUGGAUCAGA (((((((.(((((........)))))...((((.((((.......(((((...))))).((((.......))))......(((((((......)))).))).)))))))))))))))... ( -36.10) >DroYak_CAF1 11551 120 + 1 GGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAACACAUUAUAAUCCUGUUUCAAGAACAGCGGGCGUAGCUGAAUAUUGGGGCUGAAAUCUCGGUGCCCAUUAAAGCCAUGGAUCAGA (((((((.((((...((..(...((((.(((((...........)))))((((...))))...))))....)..))((.(((((((((....))))).)))).)).)))))))))))... ( -36.10) >consensus GGUUCAUAGGCUAGUUUUGGAUAGUCCAAGGAUAUCAAAUUGUAAUCCUGUGUCAGGAACAGCGGACGUAGCUGGAUAUUGGGACUGAAAUCGCGGUGCCCGUUGAAUCCAUGGAUCAGA (((((((.((.....(((((.....)))))((((((.........(((((...))))).((((.......))))))))))(((((((......)))).))).......)))))))))... (-31.74 = -32.05 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:13 2006