
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,209,846 – 5,209,955 |
| Length | 109 |
| Max. P | 0.862931 |

| Location | 5,209,846 – 5,209,955 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.89 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
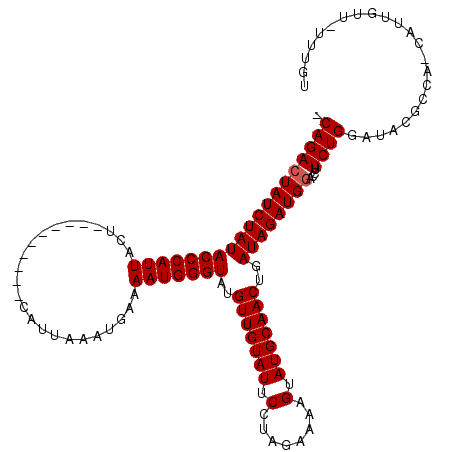
>2L_DroMel_CAF1 5209846 109 + 22407834 CCAGAGUAUCUAUACCCAUUACU----------CGUUAAAUGAAAAUGGGUAUGUUGUAUUCCUAGAAAAGUAUGCAACUGAUAGAUGGACCUUCUGAAUACGACA-CAUUGUUUUUUGU .(((((.....(((((((((..(----------(((...)))).)))))))))((((((((((((....(((.....)))..)))..((.....))))))))))).-.......))))). ( -23.70) >DroSec_CAF1 86842 115 + 1 -CAGACUAUCUAUACCCAUUACUGAAAAUUACCCAUUAAAUGAAAAUGGGUAUGUUGUAUUCCUAGAAAAGUAUGCAACAGAUAGAUGGACCUUCUGGAUACGCCAUCAUGGG----UGU -..........((((((((..........((((((((.......))))))))((((((((..((.....)).))))))))....(((((.((....)).....))))))))))----))) ( -32.70) >DroSim_CAF1 59805 110 + 1 -CAGACUAUCUAUACCCAUUACU--------UUCAUAAAAUGAAAAUGGGUAUGUUGUAUUCCCAGAAAAGUAUGCAACUGAUAGAUGGACCUUCUGGAUACGCCA-CAUUGUUCUUUGU -(((((((((.(((((((((...--------(((((...))))))))))))))(((((((.(........).))))))).)))))..((((..(.(((.....)))-.)..)))))))). ( -26.90) >consensus _CAGACUAUCUAUACCCAUUACU__________CAUUAAAUGAAAAUGGGUAUGUUGUAUUCCUAGAAAAGUAUGCAACUGAUAGAUGGACCUUCUGGAUACGCCA_CAUUGUU_UUUGU .(((((((((((((((((((........................)))))))..(((((((.(........).)))))))..))))))))....))))....................... (-21.56 = -21.89 + 0.33)

| Location | 5,209,846 – 5,209,955 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -19.78 |
| Energy contribution | -19.56 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5209846 109 - 22407834 ACAAAAAACAAUG-UGUCGUAUUCAGAAGGUCCAUCUAUCAGUUGCAUACUUUUCUAGGAAUACAACAUACCCAUUUUCAUUUAACG----------AGUAAUGGGUAUAGAUACUCUGG ........((..(-((((((((((.((((((.((.(.....).))...))))))....))))))...((((((((((((.......)----------)).))))))))).)))))..)). ( -27.00) >DroSec_CAF1 86842 115 - 1 ACA----CCCAUGAUGGCGUAUCCAGAAGGUCCAUCUAUCUGUUGCAUACUUUUCUAGGAAUACAACAUACCCAUUUUCAUUUAAUGGGUAAUUUUCAGUAAUGGGUAUAGAUAGUCUG- ..(----((((((((((.....((....)).)))))....(((((.((.(((....))).)).)))))((((((((.......))))))))..........))))))............- ( -28.00) >DroSim_CAF1 59805 110 - 1 ACAAAGAACAAUG-UGGCGUAUCCAGAAGGUCCAUCUAUCAGUUGCAUACUUUUCUGGGAAUACAACAUACCCAUUUUCAUUUUAUGAA--------AGUAAUGGGUAUAGAUAGUCUG- ....(((.....(-(((.....((....)).))))(((((.((((.((.(((....))).)).))))((((((((((((((...)))))--------...))))))))).)))))))).- ( -24.00) >consensus ACAAA_AACAAUG_UGGCGUAUCCAGAAGGUCCAUCUAUCAGUUGCAUACUUUUCUAGGAAUACAACAUACCCAUUUUCAUUUAAUG__________AGUAAUGGGUAUAGAUAGUCUG_ .......................((((..(((.........((((.((.(((....))).)).))))(((((((((........................))))))))).)))..)))). (-19.78 = -19.56 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:07 2006