| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,205,950 – 5,206,051 |
| Length | 101 |
| Max. P | 0.949020 |

| Location | 5,205,950 – 5,206,051 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.81 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
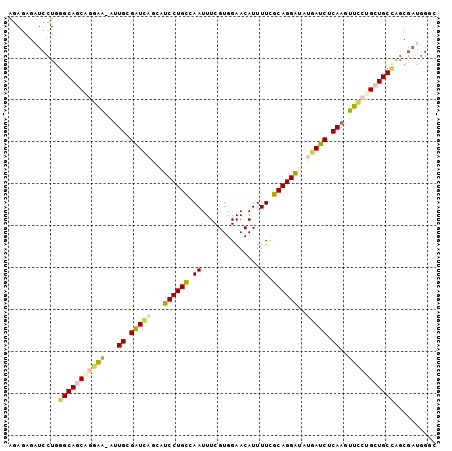
>2L_DroMel_CAF1 5205950 101 + 22407834 AGAGAGAUCCUGGGCAGCAGAAA-AUUGCGAUUAGCAUCCUGCCAAUUUCGUGGAACAUUUUCGCAGGAUAUGAUCUCAAGUUCCUGCUGCCAGCGAUGGGC ......(((((((.((((((.((-.(((.(((((..(((((((((......))(((....)))))))))).))))).))).)).)))))))))).))).... ( -36.40) >DroSec_CAF1 83007 101 + 1 AGAGAGAUCCUGGGCAUCGGGAA-AUUGCGAUCAGCAUCCUGCCAAUUUCGUGGAACAUUUUCGCAGGAUAUGAUCUCAAGUUCCUGCUGCCAGCGAUGGGC ......(((((((.((.((((((-.(((.(((((..(((((((((......))(((....)))))))))).))))).))).)))))).)))))).))).... ( -38.10) >DroSim_CAF1 56006 101 + 1 AGAGAGAUCCUGGGCAUCGGGAA-AUUGCGAUCAGCAUCCUGCCAAUUUCGUGGAACAUUUUCGCAGGAAAUGAUCUCAAGUUCCUGCUGCCAGCGAUGGGC ......(((((((.((.((((((-.(((.(((((...((((((((......))(((....)))))))))..))))).))).)))))).)))))).))).... ( -36.50) >DroEre_CAF1 85186 102 + 1 AGAGAGCUCCUGGGCAGCACGAAAAUUGCGAUCAGUAUCCUGUGAAUUUCGUGGAACAUUUUUGCAGGAUAUGAUCUCAAGUUUCCGCUGCCUGCGAUGGCC .....((((.((((((((..((((.(((.(((((.(((((((..((..............))..)))))))))))).))).)))).)))))))).))..)). ( -38.44) >DroYak_CAF1 85053 101 + 1 AGAGAGAUCCUGGGCAGCACGAA-AUUGCGAUCAGUAUCCUGCGAAUUUCGUGGAACAUUUUUGCAGGACACAAUCUCAAGUUCCCGCUGCUGGCGAUGGUC ......(((.(.((((((..(((-.(((.(((..((.(((((((((..............))))))))).)).))).))).)))..)))))).).))).... ( -31.54) >DroAna_CAF1 86149 99 + 1 AAAGAGAACUU--GCAGCAGGGG-AGUGUGUUCGGGGUCCUGCAAAUUUCGUGGAACAUUUUUGCAGGGCCAGAACACAAAUCCCUGCUGCCAUCAAAUGGC ...........--((((((((((-..(((((((..(((((((((((..............))))))))))).)))))))..))))))))))........... ( -50.44) >consensus AGAGAGAUCCUGGGCAGCAGGAA_AUUGCGAUCAGCAUCCUGCCAAUUUCGUGGAACAUUUUCGCAGGAUAUGAUCUCAAGUUCCUGCUGCCAGCGAUGGGC ............(((((((((((...((.(((((...((((((.((..............)).))))))..))))).))..))))))))))).......... (-29.38 = -29.81 + 0.43)


| Location | 5,205,950 – 5,206,051 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -29.17 |
| Energy contribution | -29.68 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.34 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
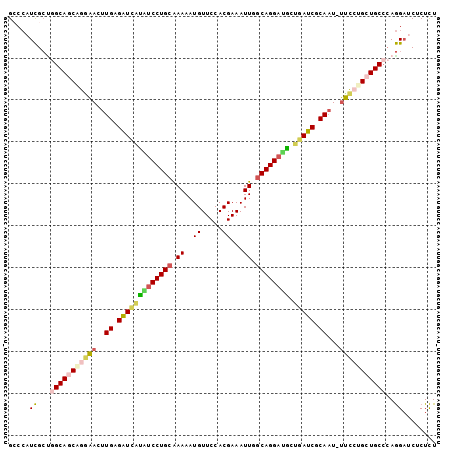
>2L_DroMel_CAF1 5205950 101 - 22407834 GCCCAUCGCUGGCAGCAGGAACUUGAGAUCAUAUCCUGCGAAAAUGUUCCACGAAAUUGGCAGGAUGCUAAUCGCAAU-UUUCUGCUGCCCAGGAUCUCUCU ....(((.(((((((((((((.(((.(((..((((((((.((..((.....))...)).))))))))...))).))).-))))))))).)))))))...... ( -38.00) >DroSec_CAF1 83007 101 - 1 GCCCAUCGCUGGCAGCAGGAACUUGAGAUCAUAUCCUGCGAAAAUGUUCCACGAAAUUGGCAGGAUGCUGAUCGCAAU-UUCCCGAUGCCCAGGAUCUCUCU ....(((.((((((.(.((((.(((.(((((((((((((.((..((.....))...)).)))))))).))))).))).-)))).).)).)))))))...... ( -35.50) >DroSim_CAF1 56006 101 - 1 GCCCAUCGCUGGCAGCAGGAACUUGAGAUCAUUUCCUGCGAAAAUGUUCCACGAAAUUGGCAGGAUGCUGAUCGCAAU-UUCCCGAUGCCCAGGAUCUCUCU ....(((.((((((.(.((((.(((.(((((..((((((.((..((.....))...)).))))))...))))).))).-)))).).)).)))))))...... ( -32.80) >DroEre_CAF1 85186 102 - 1 GGCCAUCGCAGGCAGCGGAAACUUGAGAUCAUAUCCUGCAAAAAUGUUCCACGAAAUUCACAGGAUACUGAUCGCAAUUUUCGUGCUGCCCAGGAGCUCUCU (((..((...(((((((((((.(((.((((((((((((......................))))))).))))).))).)))).)))))))..)).))).... ( -33.45) >DroYak_CAF1 85053 101 - 1 GACCAUCGCCAGCAGCGGGAACUUGAGAUUGUGUCCUGCAAAAAUGUUCCACGAAAUUCGCAGGAUACUGAUCGCAAU-UUCGUGCUGCCCAGGAUCUCUCU ........((.((((((.(((.(((.(((((((((((((....................))))))))).)))).))).-))).))))))...))........ ( -34.55) >DroAna_CAF1 86149 99 - 1 GCCAUUUGAUGGCAGCAGGGAUUUGUGUUCUGGCCCUGCAAAAAUGUUCCACGAAAUUUGCAGGACCCCGAACACACU-CCCCUGCUGC--AAGUUCUCUUU ...........(((((((((...(((((((.((.((((((((..((.....))...))))))))..)).)))))))..-.)))))))))--........... ( -42.20) >consensus GCCCAUCGCUGGCAGCAGGAACUUGAGAUCAUAUCCUGCAAAAAUGUUCCACGAAAUUGGCAGGAUGCUGAUCGCAAU_UUCCUGCUGCCCAGGAUCUCUCU .....((...(((((((((((..((.(((((((((((((.((..((.....))...)).)))))))).))))).))...)))))))))))..))........ (-29.17 = -29.68 + 0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:03 2006